7VKJ
 
 | | Structure of ESRP1 qRRM3 domain | | Descriptor: | Epithelial splicing regulatory protein 1 | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of ESRP1 qRRM3 domain
To Be Published
|
|
5EN1
 
 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*AP*CP*UP*G)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2015-11-09 | | Release date: | 2016-11-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1
Nat Commun, 2018
|
|
3REI
 
 | |
1Q1C
 
 | | Crystal structure of N(1-260) of human FKBP52 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, FK506-binding protein 4 | | Authors: | Wu, B, Li, P, Lou, Z, Ding, Y, Shu, C, Shen, B, Rao, Z. | | Deposit date: | 2003-07-18 | | Release date: | 2004-06-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3REL
 
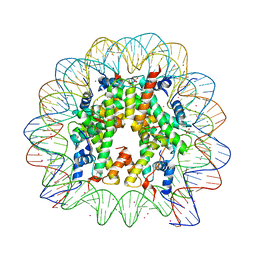 | |
3REK
 
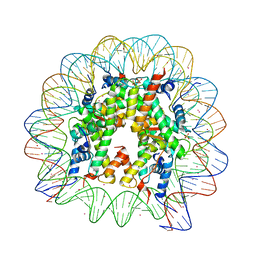 | |
5EIP
 
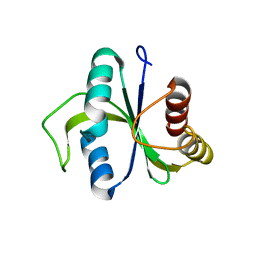 | | apo-structure of YTH domain of SpMmi1 | | Descriptor: | YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5EIM
 
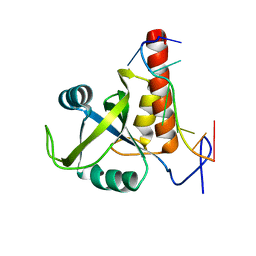 | | YTH domain-containing protein mmi1 and RNA complex | | Descriptor: | RNA (5'-R(*AP*UP*UP*AP*AP*AP*CP*A)-3'), YTH domain-containing protein mmi1 | | Authors: | Wu, B.X, Xu, J.H, Su, S.C, Ma, J.B. | | Deposit date: | 2015-10-30 | | Release date: | 2016-01-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural insights into the specific recognition of DSR by the YTH domain containing protein Mmi1
Biochem. Biophys. Res. Commun., 491, 2017
|
|
3OE6
 
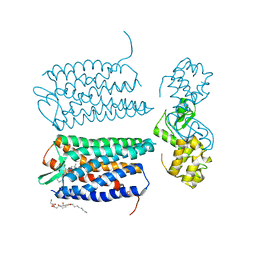 | | Crystal structure of the CXCR4 chemokine receptor in complex with a small molecule antagonist IT1t in I222 spacegroup | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (6,6-dimethyl-5,6-dihydroimidazo[2,1-b][1,3]thiazol-3-yl)methyl N,N'-dicyclohexylimidothiocarbamate, C-X-C chemokine receptor type 4, ... | | Authors: | Wu, B, Mol, C.D, Han, G.W, Katritch, V, Chien, E.Y.T, Liu, W, Cherezov, V, Stevens, R.C, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), GPCR Network (GPCR) | | Deposit date: | 2010-08-12 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists.
Science, 330, 2010
|
|
2KKX
 
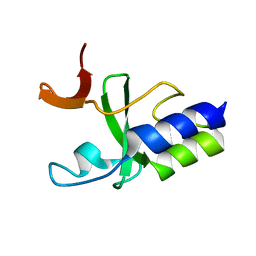 | | Solution Structure of C-terminal domain of reduced NleG2-3 (residues 90-191) from Pathogenic E. coli O157:H7. Northeast Structural Genomics Consortium and Midwest Center for Structural Genomics target ET109A | | Descriptor: | Uncharacterized protein ECs2156 | | Authors: | Wu, B, Yee, A, Fares, C, Lemak, A, Semest, A, Claude, M, Singer, A, Edwards, A, Savchenko, A, Montelione, G.T, Joachimiak, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-29 | | Release date: | 2009-08-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases.
Plos Pathog., 6, 2010
|
|
6J23
 
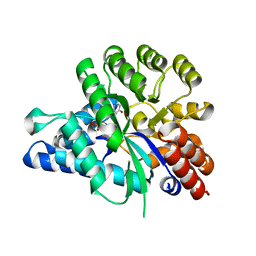 | | Crystal structure of arabidopsis ADAL complexed with GMP | | Descriptor: | Adenosine/AMP deaminase family protein, GUANOSINE-5'-MONOPHOSPHATE, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-30 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
5AC3
 
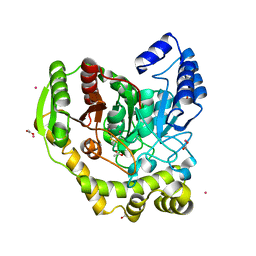 | | Crystal structure of PAM12A | | Descriptor: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | Authors: | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | Deposit date: | 2015-08-11 | | Release date: | 2016-07-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
6KIS
 
 | |
1P5Q
 
 | | Crystal Structure of FKBP52 C-terminal Domain | | Descriptor: | FK506-binding protein 4, SULFATE ION | | Authors: | Wu, B, Li, P, Lou, Z, Shu, C, Ding, Y, Shen, B, Rao, Z. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 3D structure of human FK506-binding protein 52: Implications for the assembly of the glucocorticoid receptor/Hsp90/immunophilin heterocomplex
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
2OIE
 
 | | Crystal structure of RS21-C6 core segment RSCUT | | Descriptor: | RS21-C6, SULFATE ION | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-10 | | Release date: | 2007-03-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
2OIG
 
 | | Crystal structure of RS21-C6 core segment and dm5CTP complex | | Descriptor: | 2'-DEOXY-5-METHYLCYTIDINE 5'-(TETRAHYDROGEN TRIPHOSPHATE), RS21-C6 | | Authors: | Wu, B, Liu, Y, Zhao, Q, Liao, S, Zhang, J, Bartlam, M, Chen, W, Rao, Z. | | Deposit date: | 2007-01-11 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of RS21-C6, Involved in Nucleoside Triphosphate Pyrophosphohydrolysis
J.Mol.Biol., 367, 2007
|
|
6KIT
 
 | |
6IV5
 
 | | Crystal structure of arabidopsis N6-mAMP deaminase MAPDA | | Descriptor: | Adenosine/AMP deaminase family protein, PHOSPHATE ION, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2018-12-02 | | Release date: | 2019-02-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
6KIR
 
 | | High resolution structure of mouse CXorf40A | | Descriptor: | SULFATE ION, Uncharacterized protein CXorf40 homolog | | Authors: | Wu, B.X. | | Deposit date: | 2019-07-19 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High resolution structure of mouse CXorf40A
To Be Published
|
|
7VKI
 
 | | ESRP1 qRRM2 in complex with 12mer-RNA | | Descriptor: | Epithelial splicing regulatory protein 1, RNA (12-mer) | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Nucleic Acids Res., 2023
|
|
5HO4
 
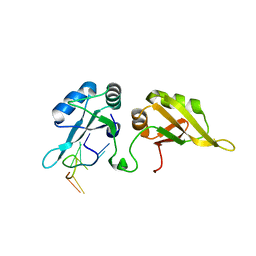 | | Crystal structure of hnRNPA2B1 in complex with 10-mer RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*AP*GP*C)-3') | | Authors: | Wu, B.X, Su, S.C, Gan, J.H, Ma, J.B. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
8IL5
 
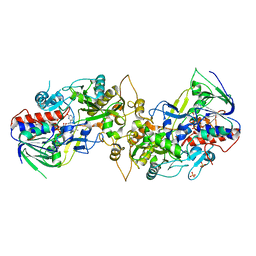 | |
2YII
 
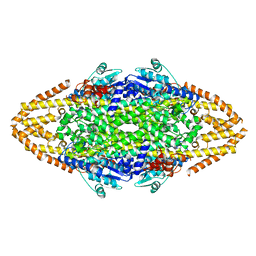 | | Manipulating the regioselectivity of phenylalanine aminomutase: new insights into the reaction mechanism of MIO-dependent enzymes from structure-guided directed evolution | | Descriptor: | BETA-MERCAPTOETHANOL, FORMIC ACID, GLYCEROL, ... | | Authors: | Wu, B, Szymanski, W, Wybenga, G.G, Heberling, M.M, Bartsch, S, Wildeman, S, Poelarends, G.J, Feringa, B.L, Dijkstra, B.W, Janssen, D.B. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism-Inspired Engineering of Phenylalanine Aminomutase for Enhanced Beta-Regioselective Asymmetric Amination of Cinnamates.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6J4T
 
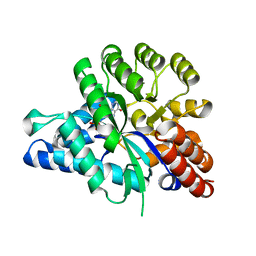 | | Crystal structure of arabidopsis ADAL complexed with IMP | | Descriptor: | Adenosine/AMP deaminase family protein, INOSINIC ACID, ZINC ION | | Authors: | Wu, B.X, Zhang, D, Nie, H.B, Shen, S.L, Li, S.S, Patel, D.J. | | Deposit date: | 2019-01-10 | | Release date: | 2019-07-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure ofArabidopsis thaliana N6-methyl-AMP deaminase ADAL with bound GMP and IMP and implications forN6-methyl-AMP recognition and processing.
Rna Biol., 16, 2019
|
|
8IL4
 
 | |
