6DFA
 
 | | Kaiso (ZBTB33) E535A zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.908 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DMX
 
 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DNQ
 
 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6DF9
 
 | | Kaiso (ZBTB33) E535Q zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.319 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DF8
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS), pH 6.5 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.536 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DF5
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DFB
 
 | | Kaiso (ZBTB33) K539A zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
6DFC
 
 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with the specific Kaiso binding sequence (KBS) with a T-to-U substitution | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*AP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*UP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2018-05-14 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conformational switch in the zinc finger protein Kaiso mediates differential readout of specific and methylated DNA sequences.
Biochemistry, 2020
|
|
1AX3
 
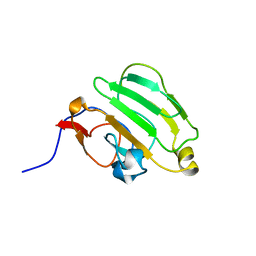 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
3QL3
 
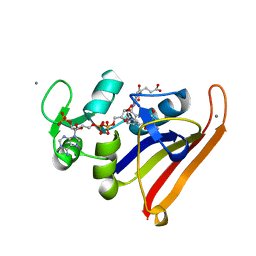 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3QL0
 
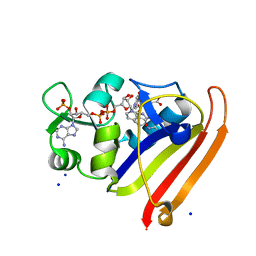 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
1TF3
 
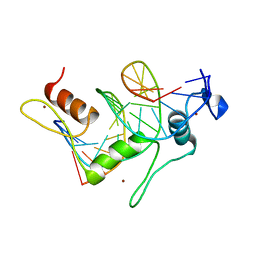 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
1NO8
 
 | | SOLUTION STRUCTURE OF THE NUCLEAR FACTOR ALY RBD DOMAIN | | Descriptor: | ALY | | Authors: | Perez-Alvarado, G.C, Martinez-Yamout, M, Allen, M.M, Grosschedl, R, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-01-15 | | Release date: | 2003-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Nuclear Factor ALY: Insights into Post-Transcriptional
Regulatory and mRNA Nuclear Export Processes
Biochemistry, 42, 2003
|
|
1POU
 
 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
1ZU1
 
 | | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | | Descriptor: | RNA binding protein ZFa, ZINC ION | | Authors: | Moller, H.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-05-29 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc fingers of the Xenopus laevis double-stranded RNA-binding protein ZFa
J.Mol.Biol., 351, 2005
|
|
1COU
 
 | |
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
1F62
 
 | | WSTF-PHD | | Descriptor: | TRANSCRIPTION FACTOR WSTF, ZINC ION | | Authors: | Pascual, J, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-19 | | Release date: | 2000-12-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor.
J.Mol.Biol., 304, 2000
|
|
1F81
 
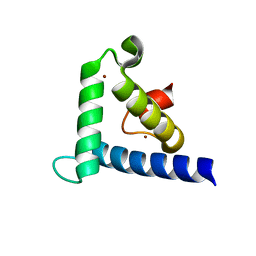 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
1G7O
 
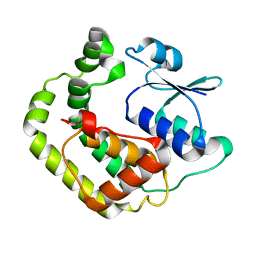 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
1DGQ
 
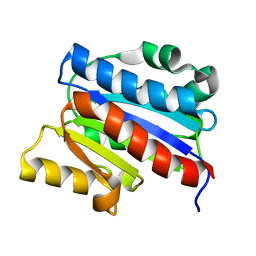 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1KBH
 
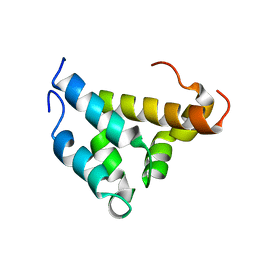 | | Mutual Synergistic Folding in the Interaction Between Nuclear Receptor Coactivators CBP and ACTR | | Descriptor: | CREB-BINDING PROTEIN, nuclear receptor coactivator | | Authors: | Demarest, S.J, Martinez-Yamout, M, Chung, J, Chen, H, Xu, W, Dyson, H.J, Evans, R.M, Wright, P.E. | | Deposit date: | 2001-11-06 | | Release date: | 2002-02-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators.
Nature, 415, 2002
|
|
1I17
 
 | | NMR STRUCTURE OF MOUSE DOPPEL 51-157 | | Descriptor: | PRION-LIKE PROTEIN | | Authors: | Mo, H, Moore, R.C, Cohen, F.E, Westaway, D, Prusiner, S.B, Wright, P.E, Dyson, H.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two different neurodegenerative diseases caused by proteins with similar structures.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1L8C
 
 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
