6WLH
 
 | |
1F68
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
1ZNF
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
7LVS
 
 | | The CBP TAZ1 Domain in Complex with a CITED2-HIF-1-Alpha Fusion Peptide | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone lysine acetyltransferase CREBBP, ZINC ION | | Authors: | Appling, F.D, Berlow, R.B, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2021-02-26 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The molecular basis of allostery in a facilitated dissociation process.
Structure, 29, 2021
|
|
1ZU1
 
 | | Solution Structure of the N-terminal Zinc Fingers of the Xenopus laevis double stranded RNA binding protein ZFa | | Descriptor: | RNA binding protein ZFa, ZINC ION | | Authors: | Moller, H.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2005-05-29 | | Release date: | 2005-09-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the N-terminal zinc fingers of the Xenopus laevis double-stranded RNA-binding protein ZFa
J.Mol.Biol., 351, 2005
|
|
1POU
 
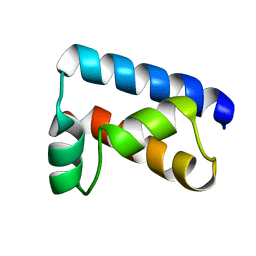 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
8T5X
 
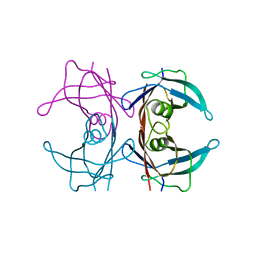 | | Probing the dissociation pathway of a kinetically labile transthyretin mutant (A25T) | | Descriptor: | Transthyretin | | Authors: | Ferguson, J.A, Sun, X, Leach, B.I, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Probing the Dissociation Pathway of a Kinetically Labile Transthyretin Mutant.
J.Am.Chem.Soc., 146, 2024
|
|
8W1N
 
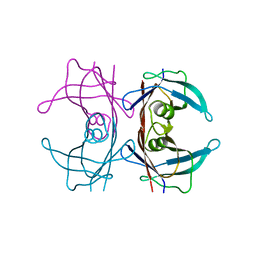 | |
8W2W
 
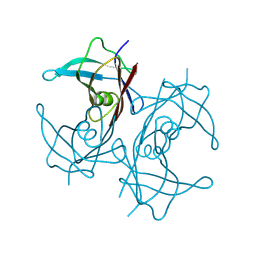 | | Structure of transthyretin synthetic mutation A120L | | Descriptor: | Transthyretin | | Authors: | Yang, K, Sun, X, Ferguson, J.A, Stanfield, R.L, Wright, P.E. | | Deposit date: | 2024-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mispacking of the F87 sidechain drives aggregation-promoting conformational fluctuations in the subunit interfaces of the transthyretin tetramer.
Protein Sci., 33, 2024
|
|
4NX7
 
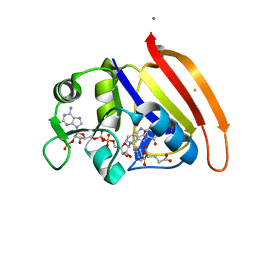 | | single cryogenic temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NX6
 
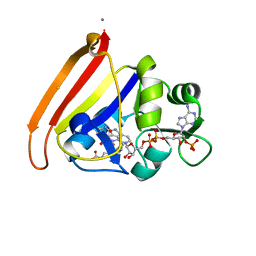 | | single room temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
2JP9
 
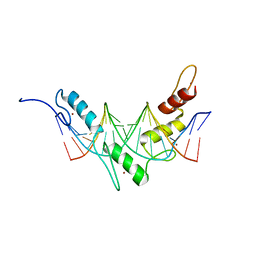 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DGP*DCP*DGP*DC)-3'), DNA (5'-D(P*DGP*DCP*DGP*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-04-30 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JOX
 
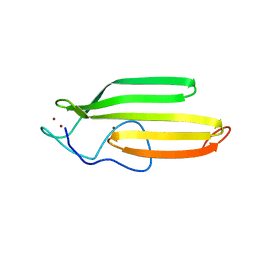 | | Embryonic Neural Inducing Factor Churchill is not a DNA-Binding Zinc Finger Protein: Solution Structure Reveals a Solvent-Exposed beta-Sheet and Zinc Binuclear Cluster | | Descriptor: | Churchill protein, ZINC ION | | Authors: | Lee, B.M, Buck-Koehntop, B.A, Martinez-Yamout, M.A, Gottesfeld, J.M, Dyson, H, Wright, P.E. | | Deposit date: | 2007-04-07 | | Release date: | 2007-08-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Embryonic Neural Inducing Factor Churchill Is not a DNA-binding Zinc Finger Protein: Solution Structure Reveals a Solvent-exposed beta-Sheet and Zinc Binuclear Cluster
J.Mol.Biol., 371, 2007
|
|
2KA4
 
 | | NMR structure of the CBP-TAZ1/STAT2-TAD complex | | Descriptor: | Crebbp protein, Signal transducer and activator of transcription 2, ZINC ION | | Authors: | Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2008-10-30 | | Release date: | 2009-04-21 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains
Embo J., 28, 2009
|
|
5HPD
 
 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HOU
 
 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HP0
 
 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4F6N
 
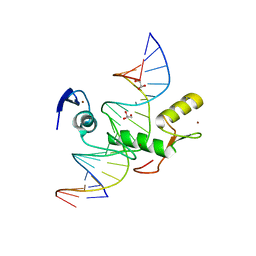 | | Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*GP*AP*(5CM)P*GP*(5CM)P*GP*GP*TP*GP*AP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*TP*CP*AP*CP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*AP*TP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F6M
 
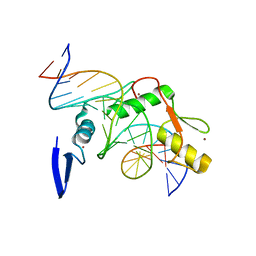 | | Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1COU
 
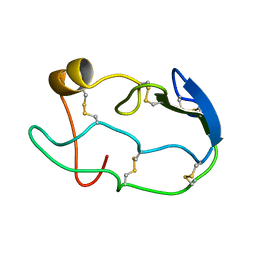 | |
1AX3
 
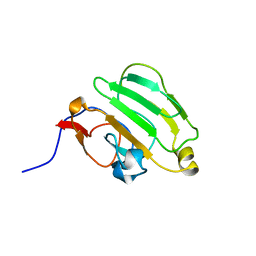 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
1KDX
 
 | | KIX DOMAIN OF MOUSE CBP (CREB BINDING PROTEIN) IN COMPLEX WITH PHOSPHORYLATED KINASE INDUCIBLE DOMAIN (PKID) OF RAT CREB (CYCLIC AMP RESPONSE ELEMENT BINDING PROTEIN), NMR 17 STRUCTURES | | Descriptor: | CBP, CREB | | Authors: | Radhakrishnan, I, Perez-Alvarado, G.C, Dyson, H.J, Wright, P.E. | | Deposit date: | 1997-09-16 | | Release date: | 1998-11-25 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions.
Cell(Cambridge,Mass.), 91, 1997
|
|
1I17
 
 | | NMR STRUCTURE OF MOUSE DOPPEL 51-157 | | Descriptor: | PRION-LIKE PROTEIN | | Authors: | Mo, H, Moore, R.C, Cohen, F.E, Westaway, D, Prusiner, S.B, Wright, P.E, Dyson, H.J. | | Deposit date: | 2001-01-31 | | Release date: | 2001-03-07 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Two different neurodegenerative diseases caused by proteins with similar structures.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
