4NX6
 
 | | single room temperature model of DHFR | | Descriptor: | BETA-MERCAPTOETHANOL, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | Fenwick, R.B, van den Bedem, H, Fraser, J.S, Wright, P.E. | | Deposit date: | 2013-12-08 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Integrated description of protein dynamics from room-temperature X-ray crystallography and NMR.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
5HP0
 
 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HPD
 
 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HOU
 
 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1EXK
 
 | | SOLUTION STRUCTURE OF THE CYSTEINE-RICH DOMAIN OF THE ESCHERICHIA COLI CHAPERONE PROTEIN DNAJ. | | Descriptor: | DNAJ PROTEIN, ZINC ION | | Authors: | Martinez-Yamout, M, Legge, G.B, Zhang, O, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-05-03 | | Release date: | 2000-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ.
J.Mol.Biol., 300, 2000
|
|
1F62
 
 | | WSTF-PHD | | Descriptor: | TRANSCRIPTION FACTOR WSTF, ZINC ION | | Authors: | Pascual, J, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-19 | | Release date: | 2000-12-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor.
J.Mol.Biol., 304, 2000
|
|
1F81
 
 | | SOLUTION STRUCTURE OF THE TAZ2 DOMAIN OF THE TRANSCRIPTIONAL ADAPTOR PROTEIN CBP | | Descriptor: | CREB-BINDING PROTEIN, ZINC ION | | Authors: | De Guzman, R.N, Liu, H.L, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2000-06-28 | | Release date: | 2000-10-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP.
J.Mol.Biol., 303, 2000
|
|
6DNQ
 
 | | HBZ77 in complex with KIX and c-Myb | | Descriptor: | 1,2-ETHANEDIOL, BZIP factor, CREB-binding protein, ... | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1L8C
 
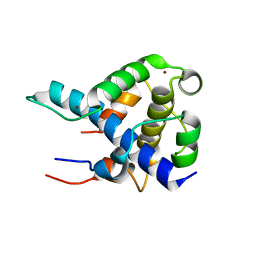 | | STRUCTURAL BASIS FOR HIF-1ALPHA/CBP RECOGNITION IN THE CELLULAR HYPOXIC RESPONSE | | Descriptor: | CREB-binding protein, Hypoxia-inducible factor 1 alpha, ZINC ION | | Authors: | Dames, S.A, Martinez-Yamout, M, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2002-03-19 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1G7O
 
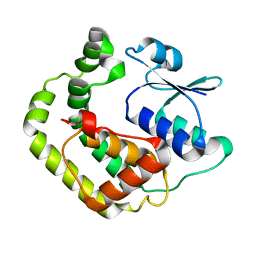 | | NMR SOLUTION STRUCTURE OF REDUCED E. COLI GLUTAREDOXIN 2 | | Descriptor: | GLUTAREDOXIN 2 | | Authors: | Xia, B, Vlamis-Gardikas, A, Holmgren, A, Wright, P.E, Dyson, H.J. | | Deposit date: | 2000-11-10 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Escherichia coli glutaredoxin-2 shows similarity to mammalian glutathione-S-transferases.
J.Mol.Biol., 310, 2001
|
|
3QL3
 
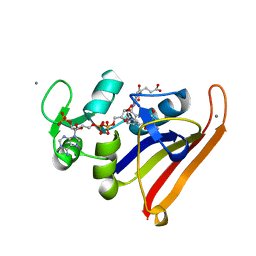 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
1IE5
 
 | | NMR STRUCTURE OF THE THIRD IMMUNOGLOBULIN DOMAIN FROM THE NEURAL CELL ADHESION MOLECULE. | | Descriptor: | NEURAL CELL ADHESION MOLECULE | | Authors: | Atkins, A.R, Chung, J, Deechongkit, S, Little, E.B, Edelman, G.M, Wright, P.E, Cunningham, B.A, Dyson, H.J. | | Deposit date: | 2001-04-06 | | Release date: | 2001-08-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third immunoglobulin domain of the neural cell adhesion molecule N-CAM: can solution studies define the mechanism of homophilic binding?
J.Mol.Biol., 311, 2001
|
|
3QL0
 
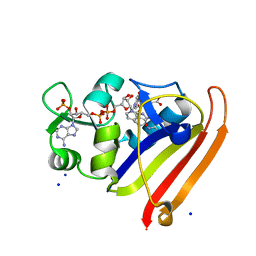 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
2HDP
 
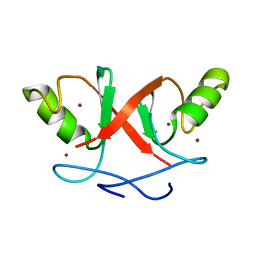 | | Solution Structure of Hdm2 RING Finger Domain | | Descriptor: | Ubiquitin-protein ligase E3 Mdm2, ZINC ION | | Authors: | Kostic, M, Matt, T, Yamout-Martinez, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2006-06-20 | | Release date: | 2006-11-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hdm2 C2H2C4 RING, a domain critical for ubiquitination of p53.
J.Mol.Biol., 363, 2006
|
|
1R8U
 
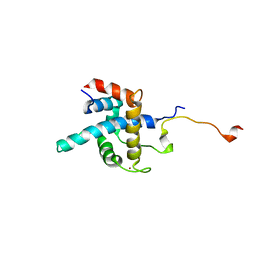 | | NMR structure of CBP TAZ1/CITED2 complex | | Descriptor: | CREB-binding protein, Cbp/p300-interacting transactivator 2, ZINC ION | | Authors: | De Guzman, R.N, Martinez-Yamout, M, Dyson, H.J, Wright, P.E. | | Deposit date: | 2003-10-28 | | Release date: | 2004-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Interaction of the TAZ1 domain of the CREB-binding protein with the activation domain of CITED2: regulation by competition between intrinsically unstructured ligands for non-identical binding sites.
J.Biol.Chem., 279, 2004
|
|
1SB0
 
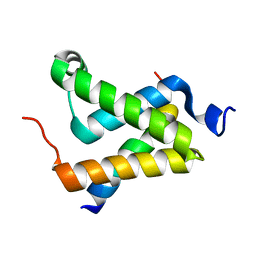 | | Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb | | Descriptor: | protein CBP, protein c-Myb | | Authors: | Zor, T, De Guzman, R.N, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-02-09 | | Release date: | 2004-04-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the KIX Domain of CBP Bound to the Transactivation
Domain of c-Myb
J.Mol.Biol., 337, 2004
|
|
5U7G
 
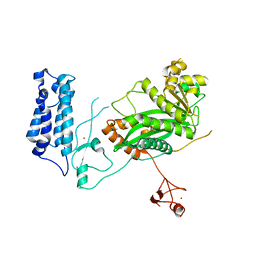 | | Crystal Structure of the Catalytic Core of CBP | | Descriptor: | CREB-binding protein, ZINC ION | | Authors: | Park, S, Stanfield, R.L, Martinez-Yamout, M.M, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2016-12-12 | | Release date: | 2017-06-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Role of the CBP catalytic core in intramolecular SUMOylation and control of histone H3 acetylation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6DMX
 
 | | HBZ56 in complex with KIX and c-Myb | | Descriptor: | BZIP factor, CREB-binding protein, Transcriptional activator Myb | | Authors: | Yang, K, Wright, P.E, Stanfield, R.L. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cooperative regulation of KIX-mediated transcription pathways by the HTLV-1 HBZ activation domain.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
1AX3
 
 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1POU
 
 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
1DGQ
 
 | | NMR SOLUTION STRUCTURE OF THE INSERTED DOMAIN OF HUMAN LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Descriptor: | LEUKOCYTE FUNCTION ASSOCIATED ANTIGEN-1 | | Authors: | Legge, G.B, Kriwacki, R.W, Chung, J, Hommel, U, Ramage, P, Case, D.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 1999-11-24 | | Release date: | 2000-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the inserted domain of human leukocyte function associated antigen-1.
J.Mol.Biol., 295, 2000
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJL
 
 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
1MYF
 
 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
