7Y5A
 
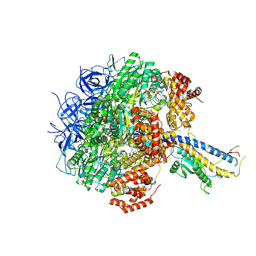 | | Cryo-EM structure of the Mycolicibacterium smegmatis F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Wong, C.F, Saw, W.-G, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
5Y4G
 
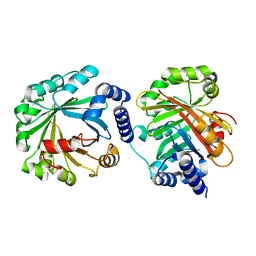 | | Apo Structure of AmbP3 | | Descriptor: | AmbP3 | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-03 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y72
 
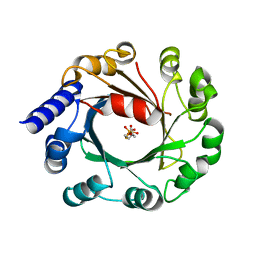 | | DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
8IYF
 
 | |
8IYE
 
 | |
5N3W
 
 | | Crystal structure of LTA4H bound to a selective inhibitor against LTB4 generation | | Descriptor: | 3-[2-(2-hydroxyphenyl)ethyl]-5-methoxy-phenol, ACETATE ION, IMIDAZOLE, ... | | Authors: | Wong, C.T, Low, C.M, Snelgrove, R.J, Hare, S.A. | | Deposit date: | 2017-02-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The development of novel LTA4H modulators to selectively target LTB4 generation.
Sci Rep, 7, 2017
|
|
5EC6
 
 | |
5EE2
 
 | |
5EE4
 
 | |
5Y84
 
 | | Hapalindole U and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole U | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-18 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
5Y7C
 
 | | Hapalindole A and DMSPP Bound AmbP3 | | Descriptor: | AmbP3, DIMETHYLALLYL S-THIOLODIPHOSPHATE, Hapalindole A | | Authors: | Wong, C.P, Awakawa, T, Nakashima, Y. | | Deposit date: | 2017-08-16 | | Release date: | 2018-07-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Two Distinct Substrate Binding Modes for the Normal and Reverse Prenylation of Hapalindoles by the Prenyltransferase AmbP3
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3GJA
 
 | | CytC3 | | Descriptor: | ACETATE ION, CytC3 | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
3GJB
 
 | | CytC3 with Fe(II) and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CytC3, ... | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
3U33
 
 | |
7OP7
 
 | | Bacteroides thetaiotaomicron mannosidase GH2 with beta-manno-configured N-alkyl cyclophellitol aziridine | | Descriptor: | (1R,2S,3R,4S,5R,6R)-5-(8-azidooctylamino)-6-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, BROMIDE ION, ... | | Authors: | McGregor, N.G.S, Beenakker, T.J.M, Kuo, C, Wong, C, Offen, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
7OP6
 
 | | Bacteroides thetaiotaomicron mannosidase GH2 with beta-manno-configured cyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, BROMIDE ION, ... | | Authors: | McGregor, N.G.S, Beenakker, T.J.M, Kuo, C, Wong, C, Offen, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2021-05-29 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
1ZP8
 
 | | HIV Protease with inhibitor AB-2 | | Descriptor: | Pol polyprotein, [1-((1S,2R)-1-BENZYL-2-HYDROXY-3-{ISOBUTYL[(4-METHOXYPHENYL)SULFONYL]AMINO}PROPYL)-1H-1,2,3-TRIAZOL-4-YL]METHYL (1R,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YLCARBAMATE | | Authors: | Brik, A, Alexandratos, J.N, Elder, J.H, Olson, A.J, Wlodawer, A, Goodsell, D.S, Wong, C.H. | | Deposit date: | 2005-05-16 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors.
Chembiochem, 6, 2005
|
|
1HOW
 
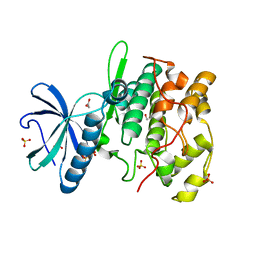 | | THE X-RAY CRYSTAL STRUCTURE OF SKY1P, AN SR PROTEIN KINASE IN YEAST | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE YMR216C, SULFATE ION | | Authors: | Nolen, B.J, Yun, C.Y, Wong, C.F, McCammon, J.A, Fu, X.-D, Ghosh, G. | | Deposit date: | 2000-12-11 | | Release date: | 2001-02-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Sky1p reveals a novel mechanism for constitutive activity.
Nat.Struct.Biol., 8, 2001
|
|
7OMS
 
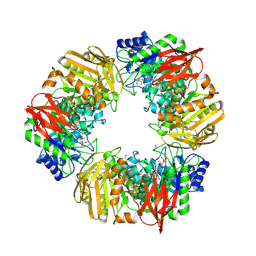 | | Bs164 in complex with mannocyclophellitol aziridine | | Descriptor: | (1~{R},2~{R},3~{S},4~{R},5~{R},6~{R})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | McGregor, N, Beenakker, T, Kuo, C.L, Wong, C.S, Offren, W.A, Armstrong, Z, Codee, J.D.C, Aerts, J.M.F.G, Florea, B.I, Overkleeft, H, Davies, G.J. | | Deposit date: | 2021-05-24 | | Release date: | 2022-02-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis of broad-specificity activity-based probes for exo -beta-mannosidases.
Org.Biomol.Chem., 20, 2022
|
|
3FWL
 
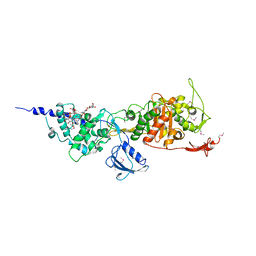 | | Crystal Structure of the Full-Length Transglycosylase PBP1b from Escherichia coli | | Descriptor: | MOENOMYCIN, Penicillin-binding protein 1B | | Authors: | Sung, M.T, Lai, Y.T, Huang, C.Y, Chou, L.Y, Wong, C.H, Ma, C. | | Deposit date: | 2009-01-19 | | Release date: | 2009-06-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.086 Å) | | Cite: | Crystal structure of the membrane-bound bifunctional transglycosylase PBP1b from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
7Y5C
 
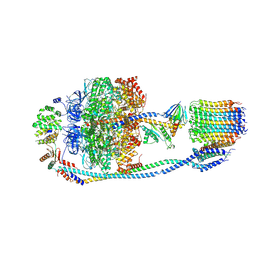 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5D
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 3) (backbone) | | Descriptor: | ATP synthase epsilon chain, ATP synthase gamma chain, ATP synthase subunit a, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7Y5B
 
 | | Cryo-EM structure of F-ATP synthase from Mycolicibacterium smegmatis (rotational state 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase epsilon chain, ... | | Authors: | Saw, W.-G, Wong, C.F, Grueber, G. | | Deposit date: | 2022-06-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural Elements Involved in ATP Hydrolysis Inhibition and ATP Synthesis of Tuberculosis and Nontuberculous Mycobacterial F-ATP Synthase Decipher New Targets for Inhibitors.
Antimicrob.Agents Chemother., 66, 2022
|
|
7XKZ
 
 | | Solution structure of subunit epsilon of the Mycobacterium abscessus F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Grueber, G, Harikishore, A, Wong, C.F, Prya, R, Dick, T. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Atomic solution structure of Mycobacterium abscessus F-ATP synthase subunit epsilon and identification of Ep1MabF1 as a targeted inhibitor.
Febs J., 289, 2022
|
|
2AKR
 
 | | Structural basis of sulfatide presentation by mouse CD1d | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zajonc, D.M, Halder, R, Wu, D, Maricic, I, Roy, K, Wong, C.-H, Kumar, V, Wilson, I.A. | | Deposit date: | 2005-08-03 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity
J.Exp.Med., 202, 2005
|
|
