6GMA
 
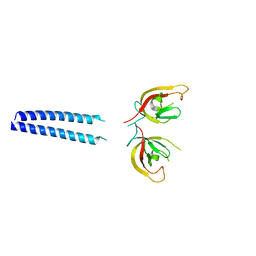 | |
2GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
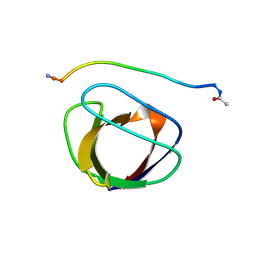
4GBQ
 
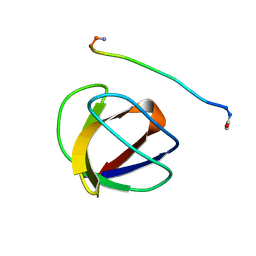 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, 15 STRUCTURES | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
3GBQ
 
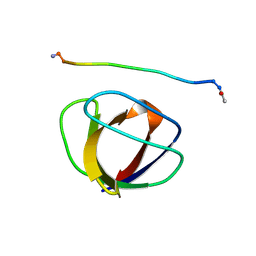 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
2K7Y
 
 | |
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1GBR
 
 | | ORIENTATION OF PEPTIDE FRAGMENTS FROM SOS PROTEINS BOUND TO THE N-TERMINAL SH3 DOMAIN OF GRB2 DETERMINED BY NMR SPECTROSCOPY | | Descriptor: | GROWTH FACTOR RECEPTOR-BOUND PROTEIN 2, SOS-A PEPTIDE | | Authors: | Wittekind, M, Mapelli, C, Farmer, B.T, Suen, K.-L, Goldfarb, V, Tsao, J, Lavoie, T, Barbacid, M, Meyers, C.A, Mueller, L. | | Deposit date: | 1994-08-12 | | Release date: | 1995-01-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy.
Biochemistry, 33, 1994
|
|
1LR0
 
 | | Pseudomonas aeruginosa TolA Domain III, Seleno-methionine Derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TolA protein, ZINC ION | | Authors: | Witty, M, Sanz, C, Shah, A, Grossman, J.G, Mizuguchi, K, Perham, R.N, Luisi, B. | | Deposit date: | 2002-05-14 | | Release date: | 2002-05-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Structure of the periplasmic domain of Pseudomonas aeruginosa TolA: evidence for an evolutionary relationship with the TonB transporter protein.
EMBO J., 21, 2002
|
|
2KLU
 
 | |
7PRM
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 13 | | Descriptor: | (4~{R})-1-[4-(4-fluorophenyl)phenyl]-4-[4-(furan-2-ylcarbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Grether, U, Gobbi, L, Kuhn, B, Collin, L, Leibrock, L, Heer, D, Wittwer, M, Benz, J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of High Brain-Penetrant and Reversible Monoacylglycerol Lipase PET Tracers for Neuroimaging.
J.Med.Chem., 65, 2022
|
|
6N64
 
 | | Crystal structure of mouse SMCHD1 hinge domain | | Descriptor: | Structural maintenance of chromosomes flexible hinge domain-containing protein 1, Uncharacterized peptide from Structural maintenance of chromosomes flexible hinge domain-containing protein 1 | | Authors: | Birkinshaw, R.W, Chen, K, Czabotar, P.E, Blewitt, M.E, Murphy, J.M. | | Deposit date: | 2018-11-25 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the hinge domain of Smchd1 reveals its dimerization mode and nucleic acid-binding residues.
Sci.Signal., 13, 2020
|
|
7BBM
 
 | | Mutant nitrobindin M75L/H76L/Q96C/M148L (NB4H) from Arabidopsis thaliana with cofactor MnPPIX | | Descriptor: | 1,2-ETHANEDIOL, MANGANESE PROTOPORPHYRIN IX, UPF0678 fatty acid-binding protein-like protein At1g79260 | | Authors: | Minges, A, Sauer, D.F, Wittwer, M, Markel, U, Spiertz, M, Schiffels, J, Davari, M.D, Okuda, J, Schwaneberg, U, Groth, G. | | Deposit date: | 2020-12-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Chemogenetic engineering of nitrobindin toward an artificial epoxygenase
Catalysis Science And Technology, 2021
|
|
4LR6
 
 | | Structure of BRD4 bromodomain 1 with a 3-methyl-4-phenylisoxazol-5-amine fragment | | Descriptor: | 3-methyl-4-phenyl-1,2-oxazol-5-amine, Bromodomain-containing protein 4, FORMIC ACID | | Authors: | Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Ravichandran, S, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
8AQF
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND LEI-515 | | Descriptor: | 1-[(~{R})-[2-chloranyl-4-[(2~{S},3~{S})-4-(3-chlorophenyl)-2,3-dimethyl-piperazin-1-yl]carbonyl-phenyl]sulfinyl]-3,3-bis(fluoranyl)pentan-2-one, Monoglyceride lipase | | Authors: | Jiang, M, Huizenga, M, Wirt, J, Paloczi, J, Amedi, A, van der Berg, R, Benz, J, Collin, L, Deng, H, Driever, W, Florea, B, Grether, U, Janssen, A, Heitman, L, Lam, T.W, Mohr, F, Pavlovic, A, Ruf, I, Rutjes, H, Stevens, F, van der Vliet, D, van der Wel, T, Wittwer, M, Boeckel, C, Pacher, P, Hohmann, A, van der Stelt, M. | | Deposit date: | 2022-08-12 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a peripheral restricted, reversible monoacylglycerol lipase inhibitor that reduces liver injury and chemotherapy-induced neuropathy
To Be Published
|
|
7ZPG
 
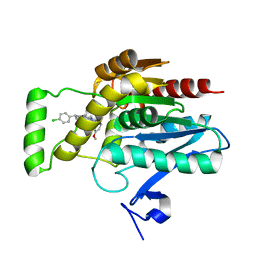 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
1MAJ
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1MAK
 
 | | SOLUTION STRUCTURE OF AN ISOLATED ANTIBODY VL DOMAIN | | Descriptor: | IGG2A-KAPPA 26-10 FV (LIGHT CHAIN) | | Authors: | Constantine, K.L, Friedrichs, M.S, Metzler, W.J, Wittekind, M, Hensley, P, Mueller, L. | | Deposit date: | 1993-09-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an isolated antibody VL domain.
J.Mol.Biol., 236, 1994
|
|
1AKP
 
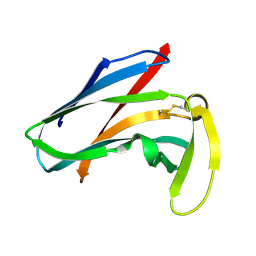 | | SEQUENTIAL 1H,13C AND 15N NMR ASSIGNMENTS AND SOLUTION CONFORMATION OF APOKEDARCIDIN | | Descriptor: | APOKEDARCIDIN | | Authors: | Constantine, K.L, Colson, K.L, Wittekind, M, Friedrichs, M.S, Zein, N, Tuttle, J, Langley, D.R, Leet, J.E, Schroeder, D.R, Lam, K.S, Farmer II, B.T, Metzler, W.J, Bruccoleri, R.E, Mueller, L. | | Deposit date: | 1994-06-20 | | Release date: | 1994-08-31 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Sequential 1H, 13C, and 15N NMR assignments and solution conformation of apokedarcidin.
Biochemistry, 33, 1994
|
|
1AW8
 
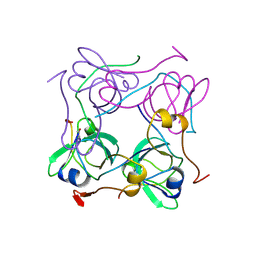 | | PYRUVOYL DEPENDENT ASPARTATE DECARBOXYLASE | | Descriptor: | L-ASPARTATE-ALPHA-DECARBOXYLASE | | Authors: | Albert, A, Dhanaraj, V, Genschel, U, Khan, G, Ramjee, M.K, Pulido, R, Sybanda, B.L, von Delf, F, Witty, M, Blundell, T.L, Smith, A.G, Abell, C. | | Deposit date: | 1997-10-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of aspartate decarboxylase at 2.2 A resolution provides evidence for an ester in protein self-processing.
Nat.Struct.Biol., 5, 1998
|
|
1PT1
 
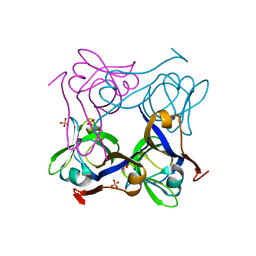 | | Unprocessed Pyruvoyl Dependent Aspartate Decarboxylase with Histidine 11 Mutated to Alanine | | Descriptor: | Aspartate 1-decarboxylase, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-22 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PQH
 
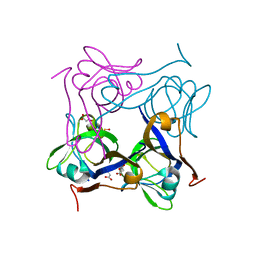 | | Serine 25 to Threonine mutation of aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase, MALONIC ACID, SODIUM ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-18 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PPY
 
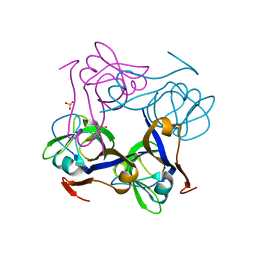 | | Native precursor of pyruvoyl dependent Aspartate decarboxylase | | Descriptor: | Aspartate 1-decarboxylase precursor, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-06-17 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1PYU
 
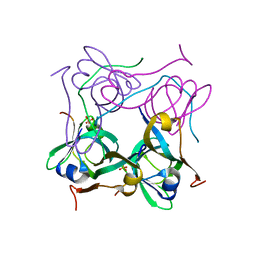 | | Processed Aspartate Decarboxylase Mutant with Ser25 mutated to Cys | | Descriptor: | Aspartate 1-decarboxylase alfa chain, Aspartate 1-decarboxylase beta chain, SULFATE ION | | Authors: | Schmitzberger, F, Kilkenny, M.L, Lobley, C.M.C, Webb, M.E, Vinkovic, M, Matak-Vinkovic, D, Witty, M, Chirgadze, D.Y, Smith, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2003-07-09 | | Release date: | 2003-11-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Constraints on protein self-processing in L-aspartate-alpha-decarboxylase
Embo J., 22, 2003
|
|
1M3U
 
 | | Crystal Structure of Ketopantoate Hydroxymethyltransferase complexed the Product Ketopantoate | | Descriptor: | 3-methyl-2-oxobutanoate hydroxymethyltransferase, KETOPANTOATE, MAGNESIUM ION | | Authors: | von Delft, F, Inoue, T, Saldanha, S.A, Ottenhof, H.H, Dhanaraj, V, Witty, M, Abell, C, Smith, A.G, Blundell, T.L. | | Deposit date: | 2002-06-30 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of E. coli Ketopantoate Hydroxymethyl Transferase Complexed with Ketopantoate and Mg(2+), Solved by Locating 160 Selenomethionine Sites.
Structure, 11, 2003
|
|
