3MG2
 
 | | Crystal structure of the orange carotenoid protein Y44S mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.653 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG1
 
 | | Crystal structure of the orange carotenoid protein from cyanobacteria Synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
3MG3
 
 | | Crystal structure of the orange carotenoid protein R155L mutant from cyanobacteria synechocystis sp. PCC 6803 | | Descriptor: | GLYCEROL, Orange carotenoid-binding protein, beta,beta-caroten-4-one | | Authors: | Wilson, A, Kinney, J, Zwart, P.H, Punginelli, C, D'Haen, S, Perreau, F, Klein, M.G, Kirilovsky, D, Kerfeld, C.A. | | Deposit date: | 2010-04-05 | | Release date: | 2010-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Structural determinants underlying photoprotection in the photoactive orange carotenoid protein of cyanobacteria.
J.Biol.Chem., 285, 2010
|
|
2FCD
 
 | | Solution structure of N-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
2FCE
 
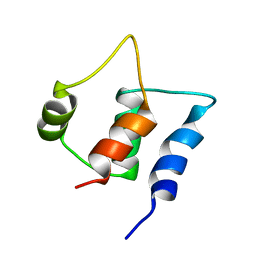 | | Solution structure of C-lobe Myosin Light Chain from Saccharomices cerevisiae | | Descriptor: | Myosin light chain 1 | | Authors: | Cicero, D.O, Pennestri, M, Contessa, G.M, Paci, M, Ragnini-Wilson, A, Melino, S. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the interaction of the myosin light chain Mlc1p with the myosin V Myo2p IQ motifs.
J.Biol.Chem., 282, 2007
|
|
7QGS
 
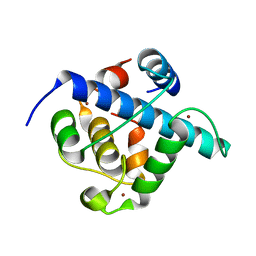 | | Crystal structure of p300 CH1 domain in complex with CITIF (a CITED2-HIF-1alpha hybrid) | | Descriptor: | Cbp/p300-interacting transactivator 2,Hypoxia-inducible factor 1-alpha, Histone acetyltransferase, ZINC ION | | Authors: | Hegedus, Z, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding p300-transcription factor interactions using sequence variation and hybridization.
Rsc Chem Biol, 3, 2022
|
|
5C3G
 
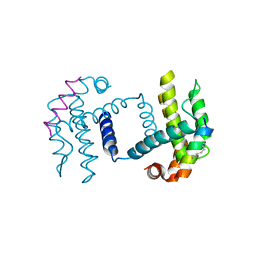 | | Crystal structure of Bcl-xl bound to BIM-MM | | Descriptor: | Bcl-2-like protein 1,Bcl-2-like protein 1, Bcl-2-like protein 11 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
5C3F
 
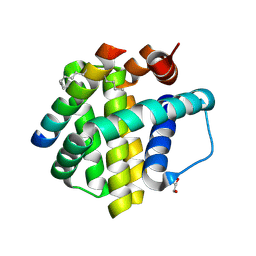 | | Crystal structure of Mcl-1 bound to BID-MM | | Descriptor: | BID-MM, GLYCEROL, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Miles, J.A, Yeo, D.J, Rowell, P, Rodriguez-Marin, S, Pask, C.M, Warriner, S.L, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Hydrocarbon constrained peptides - understanding preorganisation and binding affinity.
Chem Sci, 7, 2016
|
|
8QX5
 
 | | Helical Carotenoid Protein 4 (HCP4) from Anabaena with bound Canthaxanthin | | Descriptor: | Orange carotenoid-binding domain-containing protein, beta,beta-carotene-4,4'-dione | | Authors: | Sklyar, J, Wilson, A, Kirilovsky, D, Adir, N. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into energy quenching mechanisms and carotenoid uptake by orange carotenoid protein homologs: HCP4 and CTDH.
Int.J.Biol.Macromol., 265, 2024
|
|
8QX7
 
 | | Apo-C-Terminal Domain Homolog of the Orange Carotenoid Protein from Anabaena at a resolution of 1.95 Angstroms | | Descriptor: | All4940 protein, MALONIC ACID | | Authors: | Sklyar, J, Wilson, A, Kirilovsky, D, Adir, N. | | Deposit date: | 2023-10-22 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into energy quenching mechanisms and carotenoid uptake by orange carotenoid protein homologs: HCP4 and CTDH.
Int.J.Biol.Macromol., 265, 2024
|
|
6YR7
 
 | | 14-3-3 sigma in complex with hDMX-342+367 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Protein Mdm4 | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.105 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR5
 
 | | 14-3-3 sigma in complex with hDMX-367 peptide | | Descriptor: | 14-3-3 protein sigma, Protein Mdm4, SULFATE ION | | Authors: | Wolter, M, Srdanovic, S, Ottman, C, Warriner, S, Wilson, A. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6YR6
 
 | | 14-3-3 sigma in complex with hDM2-186 peptide | | Descriptor: | 14-3-3 protein sigma, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, ... | | Authors: | Wolter, M, Srdanovic, S, Warriner, S, Wilson, A, Ottmann, C. | | Deposit date: | 2020-04-19 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Understanding the interaction of 14-3-3 proteins with hDMX and hDM2: a structural and biophysical study.
Febs J., 2022
|
|
6HJL
 
 | | Affimer:BclxL | | Descriptor: | Affimer ADB13, Bcl-2-like protein 1 | | Authors: | Miles, J.A, Trinh, C.H, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2018-09-04 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs
Biorxiv, 2020
|
|
6STJ
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Cystatin domain-containing protein, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
6ST2
 
 | | Selective Affimers Recognize BCL-2 Family Proteins Through Non-Canonical Structural Motifs | | Descriptor: | Affimer AF6, Bcl-2-like protein 1, SULFATE ION | | Authors: | Hobor, F, Miles, J.A, Trinh, C.H, Taylor, J, Tiede, C, Rowell, P.R, Jackson, B, Nadat, F, Kyle, H.F, Wicky, B.I.M, Clarke, J, Tomlinson, D.C, Wilson, A.J, Edwards, T.A. | | Deposit date: | 2019-09-09 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Selective Affimers Recognise the BCL-2 Family Proteins BCL-x L and MCL-1 through Noncanonical Structural Motifs*.
Chembiochem, 22, 2021
|
|
8S1R
 
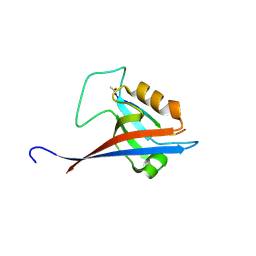 | |
7A00
 
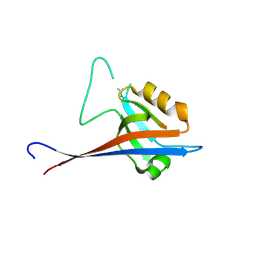 | | Crystal structure of Shank1 PDZ in complex with L6F mutant of the C-terminal hexapeptide from GKAP | | Descriptor: | L6F mutant of C-terminal hexapeptide from Guanylate kinase-associated protein, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Zsofia, H, Hetherington, K, Fruzsina, H, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-07-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Query-guided protein-protein interaction inhibitor discovery.
Chem Sci, 12, 2021
|
|
6YX0
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[2-(4-oxidanylidenebutanoyl)hydrazinyl]methyl]benzoic acid, PWQ-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6YX2
 
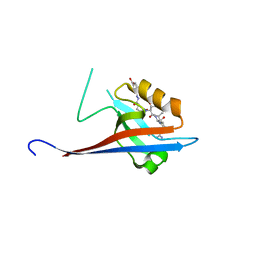 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 4-[[(~{E})-5-oxidanylidenepentanoyldiazenyl]methyl]benzoic acid, PWW-THR-ARG-LEU, SH3 and multiple ankyrin repeat domains protein 1 | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6YX1
 
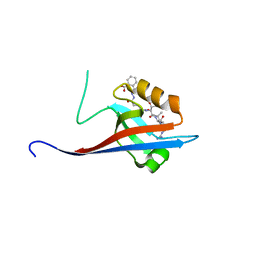 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[[2-(5-oxidanylidenepentanoyl)hydrazinyl]methyl]benzoic acid, ARGININE, LEUCINE, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6FEJ
 
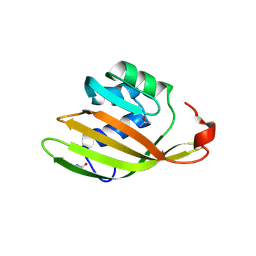 | | Anabaena Apo-C-Terminal Domain Homolog Protein | | Descriptor: | All4940 protein, UREA | | Authors: | Harris, D, Wilson, A, Muzzopappa, F, Kirilovsky, D, Adir, N. | | Deposit date: | 2018-01-02 | | Release date: | 2018-07-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural rearrangements in the C-terminal domain homolog of Orange Carotenoid Protein are crucial for carotenoid transfer.
Commun Biol, 1, 2018
|
|
6YWZ
 
 | | Crystal structure of SHANK1 PDZ in complex with a peptide-small molecule hybrid | | Descriptor: | 2-[(~{E})-(4-oxidanylidenebutanoylhydrazinylidene)methyl]benzoic acid, ARGININE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hegedus, Z, Hobor, F, Shoemark, D.K, Celis, S, Lian, L.J, Trinh, C.H, Sessions, R.B, Edwards, T.A, Wilson, A.J. | | Deposit date: | 2020-04-30 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Identification of beta-strand mediated protein-protein interaction inhibitors using ligand-directed fragment ligation.
Chem Sci, 12, 2021
|
|
6D0N
 
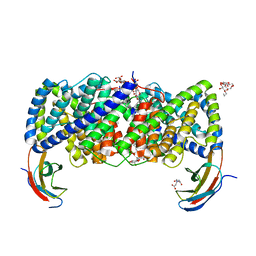 | | Crystal structure of a CLC-type fluoride/proton antiporter, V319G mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D0K
 
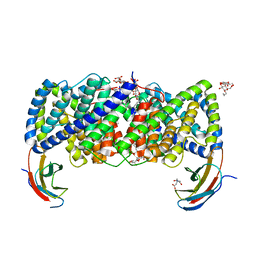 | | Crystal structure of a CLC-type fluoride/proton antiporter, E118Q mutant | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, CLC-type fluoride/proton antiporter, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | Authors: | Last, N.B, Stockbridge, R.B, Wilson, A.E, Shane, T, Kolmakova-Partensky, L, Koide, A, Koide, S, Miller, C. | | Deposit date: | 2018-04-10 | | Release date: | 2018-07-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | A CLC-type F-/H+antiporter in ion-swapped conformations.
Nat. Struct. Mol. Biol., 25, 2018
|
|
