6ZQ7
 
 | | Crystal structure of Chaetomium thermophilum Glycerol Kinase in I222 space group | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycerol kinase-like protein | | Authors: | Wilk, P, Wator, E, Grudnik, P. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.421 Å) | | Cite: | Structural Characterization of Glycerol Kinase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 21, 2020
|
|
6ZQ6
 
 | |
6F6Z
 
 | | Mouse Thymidylate Synthase Cocrystallized with N(4)OHdCMP and Soaked in Methylenetetrahydrofolate | | Descriptor: | (2~{S})-2-[[4-[[(6~{R})-2-azanyl-4-oxidanylidene-5,6,7,8-tetrahydro-1~{H}-pteridin-6-yl]methyl-methyl-amino]phenyl]carbonylamino]pentanedioic acid, 2'-deoxy-N-hydroxycytidine 5'-(dihydrogen phosphate), Thymidylate synthase | | Authors: | Wilk, P, Jarmula, A, Maj, P, Rode, W. | | Deposit date: | 2017-12-06 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.127 Å) | | Cite: | Molecular Mechanism of Thymidylate Synthase Inhibition by N4-Hydroxy-dCMP in View of Spectrophotometric and Crystallographic Studies
Int J Mol Sci, 22, 2021
|
|
6TXH
 
 | | Crystal structure of thermotoga maritima Ferritin in apo form | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXL
 
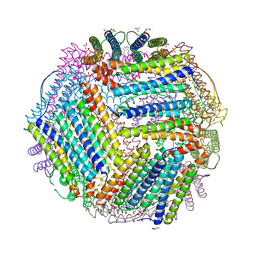 | | Crystal structure of thermotoga maritima E65Q Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXN
 
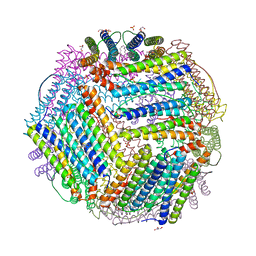 | | Crystal structure of thermotoga maritima Ferritin in apo form | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXM
 
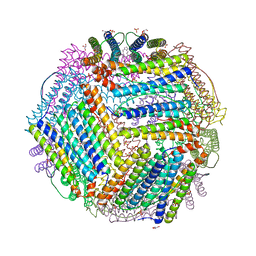 | | Crystal structure of thermotoga maritima E65R Ferritin | | Descriptor: | EICOSANE, Ferritin, GLYCEROL, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXJ
 
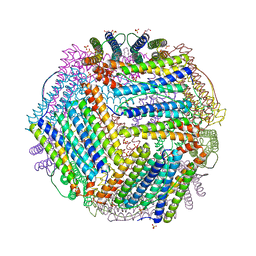 | | Crystal structure of thermotoga maritima A42V E65D Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S, Biela, A.P. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXK
 
 | | Crystal structure of thermotoga maritima E65K Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.359 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6TXI
 
 | | Crystal structure of thermotoga maritima E65A Ferritin | | Descriptor: | EICOSANE, FE (III) ION, Ferritin, ... | | Authors: | Wilk, P, Grudnik, P, Kumar, M, Heddle, J, Chakraborti, S. | | Deposit date: | 2020-01-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.759 Å) | | Cite: | A single residue can modulate nanocage assembly in salt dependent ferritin.
Nanoscale, 13, 2021
|
|
6QSC
 
 | | Crystal Structure of Arg470His mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Wator, E, Weiss, M.S. | | Deposit date: | 2019-02-20 | | Release date: | 2020-03-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.569 Å) | | Cite: | Structural analysis of new compound heterozygous variants in PEPD gene identified in a patient with Prolidase Deficiency diagnosed by exome sequencing.
Genet Mol Biol, 44, 2021
|
|
6QSB
 
 | |
4IQB
 
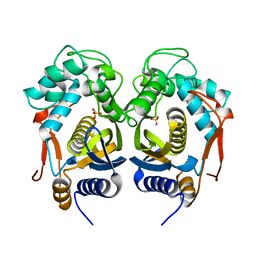 | | High Resolution Crystal Structure of C.elegans Thymidylate Synthase | | Descriptor: | SULFATE ION, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-11 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Crystal structures of nematode (parasitic T. spiralis and free living C. elegans), compared to mammalian, thymidylate synthases (TS). Molecular docking and molecular dynamics simulations in search for nematode-specific inhibitors of TS.
J. Mol. Graph. Model., 77, 2017
|
|
4IRR
 
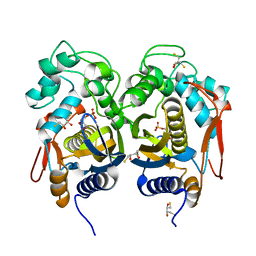 | | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-15 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal Structure of C.elegans Thymidylate Synthase in Complex with dUMP
To be Published
|
|
4ISW
 
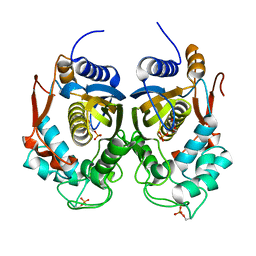 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
7B9P
 
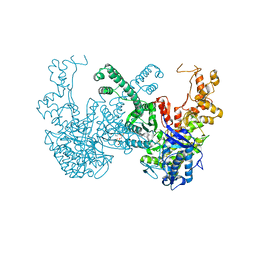 | | Structure of Ribonucleotide reductase from Rhodobacter sphaeroides | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, MAGNESIUM ION, Vitamin B12-dependent ribonucleotide reductase | | Authors: | Wilk, P, Feiler, C, Loderer, C, Kabinger, F. | | Deposit date: | 2020-12-14 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.646 Å) | | Cite: | HUG Domain Is Responsible for Active Dimer Stabilization in an NrdJd Ribonucleotide Reductase.
Biochemistry, 61, 2022
|
|
6H2Q
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and LeuPro ligand | | Descriptor: | GLYCEROL, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Weiss, M.S. | | Deposit date: | 2018-07-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
6H2P
 
 | | Crystal Structure of Arg184Gln mutant of Human Prolidase with Mn ions and Cacodylate ligand | | Descriptor: | CACODYLATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Wilk, P, Piwowarczyk, R, Weiss, M.S. | | Deposit date: | 2018-07-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
7PQF
 
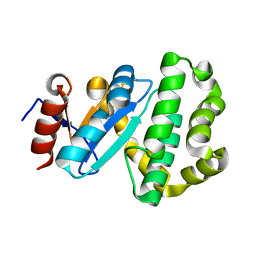 | | Crystal structure of Campylobacter jejuni DsbA2 | | Descriptor: | Thiol:disulfide interchange protein DsbA/DsbL | | Authors: | Wilk, P, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
4PSG
 
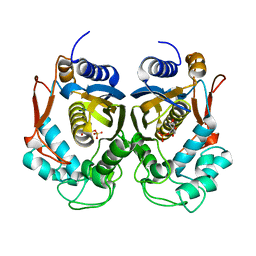 | |
7PQ7
 
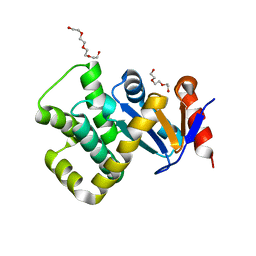 | | Crystal structure of Campylobacter jejuni DsbA1 | | Descriptor: | TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, Thiol:disulfide interchange protein DsbA | | Authors: | Wilk, P, Orlikowska, M, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-16 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
6ZQ5
 
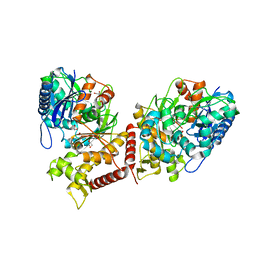 | | Crystal structure of Chaetomium thermophilum Glycerol Kinase in P2221 space group | | Descriptor: | 1,2-ETHANEDIOL, Glycerol kinase-like protein | | Authors: | Wilk, P, Wator, E, Malecki, P, Grudnik, P. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural Characterization of Glycerol Kinase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 21, 2020
|
|
6ZQ4
 
 | |
6ZQ8
 
 | | Crystal structure of Chaetomium thermophilum Glycerol Kinase in P3221 space group | | Descriptor: | Glycerol kinase-like protein | | Authors: | Wilk, P, Wator, E, Malecki, P, Tokarz, P, Grudnik, P. | | Deposit date: | 2020-07-09 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural Characterization of Glycerol Kinase from the Thermophilic Fungus Chaetomium thermophilum .
Int J Mol Sci, 21, 2020
|
|
5MC4
 
 | | Crystal Structure of Gly448Arg mutant of Human Prolidase with Mn ions and GlyPro ligand | | Descriptor: | GLYCEROL, GLYCINE, HYDROXIDE ION, ... | | Authors: | Wilk, P, Mueller, U, Dobbek, H, Weiss, M.S. | | Deposit date: | 2016-11-09 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for prolidase deficiency disease mechanisms.
FEBS J., 285, 2018
|
|
