1J01
 
 | | Crystal Structure Of The Xylanase Cex With Xylobiose-Derived Inhibitor Isofagomine lactam | | Descriptor: | (3S,4R)-3-hydroxy-2-oxopiperidin-4-yl beta-D-xylopyranoside, beta-1,4-xylanase | | Authors: | Williams, S.J, Notenboom, V, Wicki, J, Rose, D.R, Withers, S.G. | | Deposit date: | 2002-10-25 | | Release date: | 2002-11-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A New, Simple, High-Affinity Glycosidase Inhibitor: Analysis of Binding through X-ray Crystallography, Mutagenesis, and Kinetic Analysis
J.Am.Chem.Soc., 122, 2000
|
|
1V0M
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-deoxynojirimycin at pH 7.5 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, PIPERIDINE-3,4,5-TRIOL, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0N
 
 | | Xylanase Xyn10a from Streptomyces lividans in complex with xylobio-isofagomine at pH 7.5 | | Descriptor: | 1,2-ETHANEDIOL, ENDO-1,4-BETA-XYLANASE A, IMIDAZOLE, ... | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
1V0L
 
 | | Xylanase Xyn10A from Streptomyces lividans in complex with xylobio-isofagomine at pH 5.8 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, PIPERIDINE-3,4-DIOL, beta-D-xylopyranose | | Authors: | Gloster, T.M, Williams, S.J, Roberts, S, Tarling, C.A, Wicki, J, Withers, S.G, Davies, G.J. | | Deposit date: | 2004-03-31 | | Release date: | 2004-08-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic Resolution Analyses of the Binding of Xylobiose-Derived Deoxynojirimycin and Isofagomine to Xylanase Xyn10A
Chem.Commun.(Camb.), 16, 2004
|
|
8SUV
 
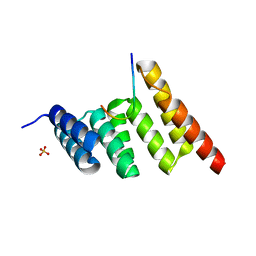 | | CHIP-TPR in complex with the C-terminus of CHIC2 | | Descriptor: | Cysteine-rich hydrophobic domain-containing protein 2, E3 ubiquitin-protein ligase CHIP, SULFATE ION | | Authors: | Cupo, A.R, McDermott, L.E, DeSilva, A.R, Callahan, M, Nix, J.C, Gestwicki, J.E, Page, R.C. | | Deposit date: | 2023-05-13 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Interaction with the membrane-anchored protein CHIC2 constrains the ubiquitin ligase activity of CHIP
Biorxiv, 2023
|
|
6XMJ
 
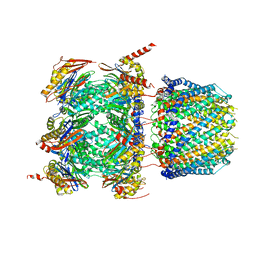 | | Human 20S proteasome bound to an engineered 11S (PA26) activator | | Descriptor: | Proteasome activator protein PA26, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | de la Pena, A.H, Opoku-Nsiah, K.A, Williams, S.K, Chopra, N, Sali, A, Gestwicki, J.E, Lander, G.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The Y Phi motif defines the structure-activity relationships of human 20S proteasome activators.
Nat Commun, 13, 2022
|
|
4ZPJ
 
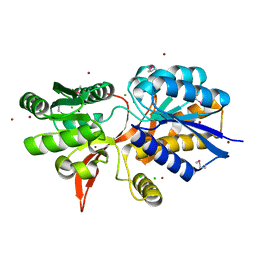 | | ABC transporter substrate-binding protein from Sphaerobacter thermophilus | | Descriptor: | CHLORIDE ION, Extracellular ligand-binding receptor, ZINC ION | | Authors: | OSIPIUK, J, Holowicki, J, Clancy, S, JOACHIMIAK, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-05-07 | | Release date: | 2015-05-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | ABC transporter substrate-binding protein from Sphaerobacter thermophilus.
to be published
|
|
6NSV
 
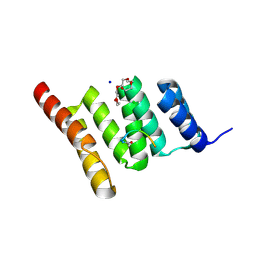 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated optimized peptide | | Descriptor: | ACE-LEU-TRP-TRP-PRO-ASP, CHLORIDE ION, E3 ubiquitin-protein ligase CHIP, ... | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2019-01-25 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.305 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
4Y7D
 
 | | Alpha/beta hydrolase fold protein from Nakamurella multipartita | | Descriptor: | Alpha/beta hydrolase fold protein, CHLORIDE ION, SODIUM ION | | Authors: | Cuff, M.E, OSIPIUK, J, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-02-14 | | Release date: | 2015-02-25 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Alpha/beta hydrolase fold protein from Nakamurella multipartita.
to be published
|
|
4XLT
 
 | | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053 | | Descriptor: | Response regulator receiver protein | | Authors: | Chang, C, Cuff, M, Holowicki, J, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-13 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of response regulator receiver protein from Dyadobacter fermentans DSM 18053
To Be Published
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7M
 
 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7O
 
 | | ATP- and mtHsp10-bound mtHsp60 V72I focus | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7N
 
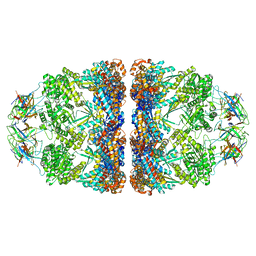 | | ATP- and mtHsp10-bound mtHsp60 V72I | | Descriptor: | 10 kDa heat shock protein, mitochondrial, 60 kDa heat shock protein, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
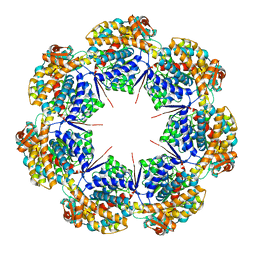 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
4KVH
 
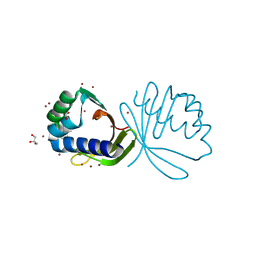 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | BROMIDE ION, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
8FYU
 
 | | Crystal structure of the human CHIP-TPR domain in complex with a 10mer acetylated tau peptide | | Descriptor: | ACE-SER-SER-THR-GLY-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-01-26 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.84839141 Å) | | Cite: | Phosphorylation of tau at a single residue inhibits binding to the E3 ubiquitin ligase, CHIP.
Nat Commun, 15, 2024
|
|
4DQ1
 
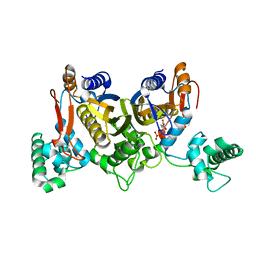 | | Thymidylate synthase from Staphylococcus aureus. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Osipiuk, J, Holowicki, J, Jedrzejczak, R, Rubin, E, Guinn, K, Ioerger, T, Baker, D, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-14 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Thymidylate synthase from Staphylococcus aureus.
To be Published
|
|
8GCK
 
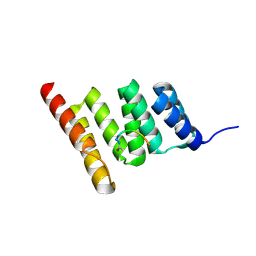 | | Crystal structure of the human CHIP-TPR domain in complex with a 6mer acetylated tau peptide | | Descriptor: | ACE-SER-ILE-ASP-MET-VAL-ASP, E3 ubiquitin-protein ligase CHIP | | Authors: | Wucherer, K, Bohn, M.F, Basu, K, Nadel, C.M, Gestwicki, J.E, Craik, C.S. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.36823535 Å) | | Cite: | Intersecting PTMs regulate clearance of pathogenic tau by the ubiquitin ligase CHIP.
To Be Published
|
|
4ML9
 
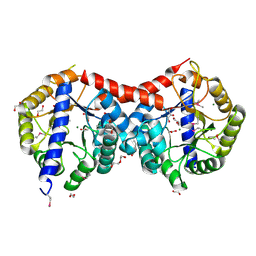 | | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kim, Y, Holowicki, J, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-06 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Crystal Structure of Uncharacterized TIM Barrel Protein with the Conserved Phosphate Binding Site fromSebaldella termitidis
To be Published
|
|
4MJD
 
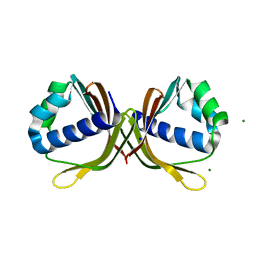 | | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747 | | Descriptor: | Ketosteroid isomerase fold protein Hmuk_0747, MAGNESIUM ION, SODIUM ION | | Authors: | Chang, C, Holowicki, J, Bearden, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-09-03 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of ketosteroid isomerase fold protein Hmuk_0747
To be Published
|
|
4GBJ
 
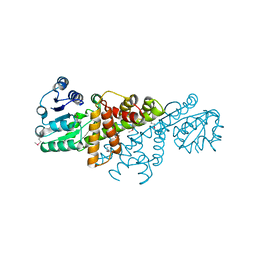 | |
4HB7
 
 | | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50. | | Descriptor: | 1,2-ETHANEDIOL, Dihydropteroate synthase | | Authors: | Cuff, M.E, Holowicki, J, Jedrzejczak, R, Terwilliger, T.C, Rubin, E.J, Guinn, K, Baker, D, Ioerger, T.R, Sacchettini, J.C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-09-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Structure of Dihydropteroate Synthase from Staphylococcus aureus subsp. aureus Mu50.
TO BE PUBLISHED
|
|
