3TEK
 
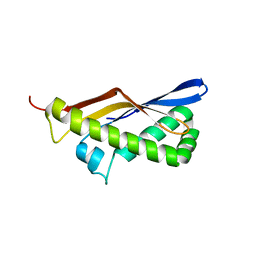 | | ThermoDBP: a non-canonical single-stranded DNA binding protein with a novel structure and mechanism | | Descriptor: | ThermoDBP-single stranded DNA binding protein | | Authors: | White, M.F, Paytubi, S, Liu, H, Graham, S, McMahon, S.A, Naismith, J.H. | | Deposit date: | 2011-08-15 | | Release date: | 2011-11-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Displacement of the canonical single-stranded DNA-binding protein in the Thermoproteales.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3ZFV
 
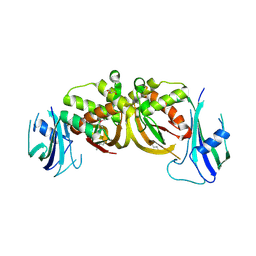 | | Crystal structure of an archaeal CRISPR-associated Cas6 nuclease | | Descriptor: | CRISPR-ASSOCIATED ENDORIBONUCLEASE CAS6 1, GLYCEROL | | Authors: | Reeks, J, Liu, H, White, M.F, Naismith, J.H. | | Deposit date: | 2012-12-12 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a Dimeric Crenarchaeal Cas6 Enzyme with an Atypical Active Site for Crispr RNA Processing
Biochem.J., 452, 2013
|
|
7QQK
 
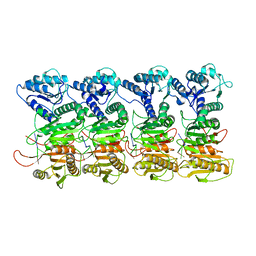 | | TIR-SAVED effector bound to cA3 | | Descriptor: | RNA (5'-R(P*AP*AP*A)-3'), TIR_SAVED fusion protein | | Authors: | Spagnolo, L, White, M.F, Hogrel, G, Guild, A. | | Deposit date: | 2022-01-09 | | Release date: | 2022-06-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cyclic nucleotide-induced helical structure activates a TIR immune effector.
Nature, 608, 2022
|
|
1HH1
 
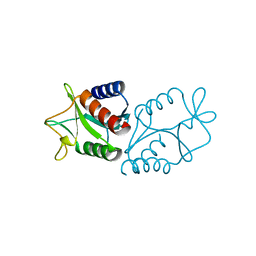 | | THE STRUCTURE OF HJC, A HOLLIDAY JUNCTION RESOLVING ENZYME FROM SULFOLOBUS SOLFATARICUS | | Descriptor: | HOLLIDAY JUNCTION RESOLVING ENZYME HJC | | Authors: | Bond, C.S, Kvaratskhelia, M, Richard, D, White, M.F, Hunter, W.N. | | Deposit date: | 2000-12-18 | | Release date: | 2001-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of Hjc, a Holliday Junction Resolvase, from Sulfolobus Solfataricus
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
8QJK
 
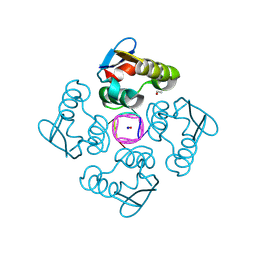 | | Structure of the cytoplasmic domain of csx23 from Vibrio cholera in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | ACETYL GROUP, Cyclic tetraadenosine monophosphate (cA4), SODIUM ION, ... | | Authors: | McMahon, S.A, McQuarrie, S, Gloster, T.M, Gruschow, S, White, M.F. | | Deposit date: | 2023-09-13 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.761 Å) | | Cite: | A cyclic-nucleotide binding membrane protein provides CRISPR-mediated antiphage defence in Vibrio cholera
To Be Published
|
|
8ANE
 
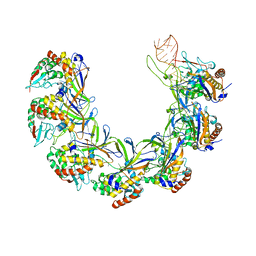 | | Structure of the type I-G CRISPR effector | | Descriptor: | Cas7, RNA (66-MER) | | Authors: | Shangguan, Q, Graham, S, Sundaramoorthy, R, White, M.F. | | Deposit date: | 2022-08-05 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and mechanism of the type I-G CRISPR effector.
Nucleic Acids Res., 50, 2022
|
|
8PCW
 
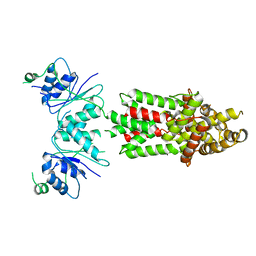 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
8PE3
 
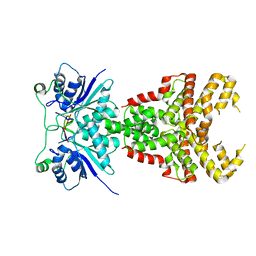 | | Structure of Csm6' from Streptococcus thermophilus in complex with cyclic hexa-adenylate (cA6) | | Descriptor: | CRISPR system endoribonuclease Csm6', Cyclic hexaadenosine monophosphate (cA6), RNA | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-13 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
6YUD
 
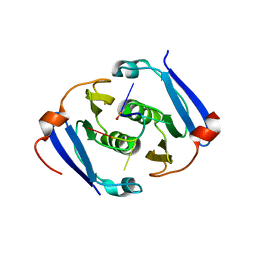 | | Structure of Csx3/Crn3 from Archaeoglobus fulgidus in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Cyclic tetraadenosine monophosphate (cA4), Uncharacterized protein AF_1864 | | Authors: | McQuarrie, S, Gloster, T.M, White, M.F, Graham, S, Athukoralage, J.S, Gruschow, S. | | Deposit date: | 2020-04-27 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.
Elife, 9, 2020
|
|
8B2X
 
 | |
3FFE
 
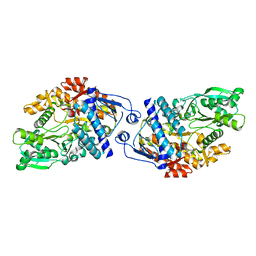 | | Structure of Achromobactin Synthetase Protein D, (AcsD) | | Descriptor: | AcsD | | Authors: | McMahon, S.A, Liu, H, Carter, L, Oke, M, Johnson, K.A, Schmelz, S, Challis, G.L, White, M.F, Naismith, J.H, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2008-12-03 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis
Nat.Chem.Biol., 5, 2009
|
|
8BMW
 
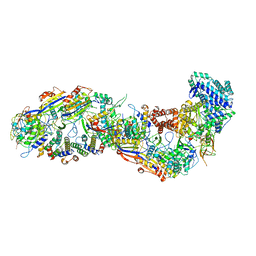 | | SsoCsm | | Descriptor: | CRISPR-associated Cas7 paralog (Type III-D), CRISPR-associated protein Cas10 (Type III-D), CRISPR-associated protein Cas5 (Type III-D), ... | | Authors: | Spagnolo, L, White, M.F. | | Deposit date: | 2022-11-11 | | Release date: | 2023-03-01 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Saccharolobus solfataricus type III-D CRISPR effector.
Curr Res Struct Biol, 5, 2023
|
|
6SCE
 
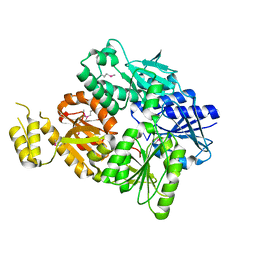 | | Structure of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Zhu, W, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate.
Nat Commun, 11, 2020
|
|
6SCF
 
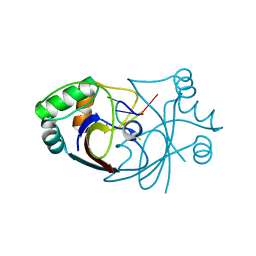 | | A viral anti-CRISPR subverts type III CRISPR immunity by rapid degradation of cyclic oligoadenylate | | Descriptor: | Uncharacterized protein, cyclic oligoadenylate | | Authors: | McMahon, S.A, Athukoralage, J.S, Graham, S, White, M.F, Gloster, T.M. | | Deposit date: | 2019-07-24 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity.
Nature, 577, 2020
|
|
2MNA
 
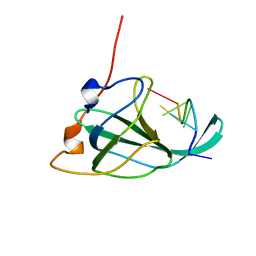 | | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus | | Descriptor: | Single-stranded DNA binding protein (SSB), ssDNA | | Authors: | Gamsjaeger, R, Kariawasam, R, Gimenez, A.X, Touma, C.F, McIlwain, E, Bernardo, R.E, Shepherd, N.E, Ataide, S.F, Dong, A.Q, Richard, D.J, White, M.F, Cubeddu, L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The structural basis of DNA binding by the single-stranded DNA-binding protein from Sulfolobus solfataricus
Biochem.J., 465, 2015
|
|
2IVY
 
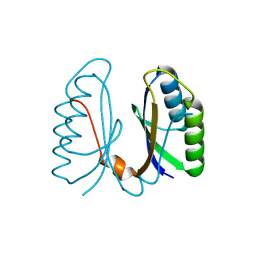 | | Crystal structure of hypothetical protein sso1404 from Sulfolobus solfataricus P2 | | Descriptor: | HYPOTHETICAL PROTEIN SSO1404 | | Authors: | Yan, X, Carter, L.G, Dorward, M, Liu, H, McMahon, S.A, Oke, M, Powers, H, White, M.F, Naismith, J.H. | | Deposit date: | 2006-06-22 | | Release date: | 2006-06-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genomics, 11, 2010
|
|
2IX2
 
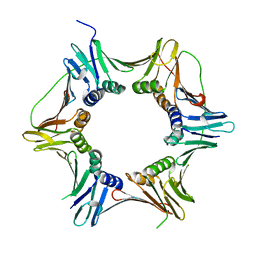 | | Crystal structure of the heterotrimeric PCNA from Sulfolobus solfataricus | | Descriptor: | DNA POLYMERASE SLIDING CLAMP A, DNA POLYMERASE SLIDING CLAMP B, DNA POLYMERASE SLIDING CLAMP C | | Authors: | Williams, G.J, Johnson, K, McMahon, S.A, Carter, L, Oke, M, Liu, H, Taylor, G.L, White, M.F, Naismith, J.H. | | Deposit date: | 2006-07-05 | | Release date: | 2006-10-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the Heterotrimeric PCNA from Sulfolobus Solfataricus.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
7BDV
 
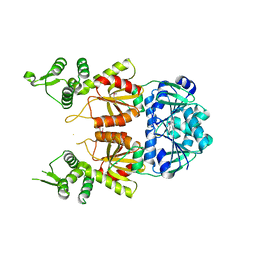 | | Structure of Can2 from Sulfobacillus thermosulfidooxidans in complex with cyclic tetra-adenylate (cA4) | | Descriptor: | Can2, Cyclic tetraadenosine monophosphate (cA4) | | Authors: | McQuarrie, S, McMahon, S.A, Gloster, T.M, White, M.F, Graham, S, Zhu, W, Gruschow, S. | | Deposit date: | 2020-12-22 | | Release date: | 2021-03-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The CRISPR ancillary effector Can2 is a dual-specificity nuclease potentiating type III CRISPR defence.
Nucleic Acids Res., 49, 2021
|
|
2BGW
 
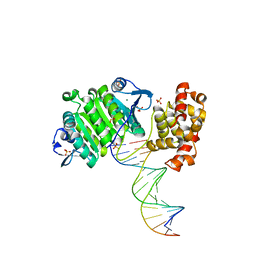 | | XPF from Aeropyrum pernix, complex with DNA | | Descriptor: | 5'-D(*GP*AP*TP*CP*AP*CP*AP*GP*AP*TP *GP*CP*TP*GP*A)-3', 5'-D(*TP*CP*AP*GP*CP*AP*TP*CP*TP*GP *TP*GP*AP*TP*C)-3', MAGNESIUM ION, ... | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-06 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BKY
 
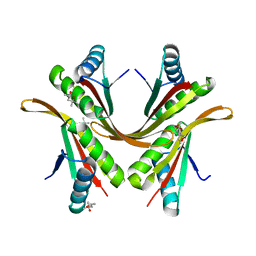 | | Crystal structure of the Alba1:Alba2 heterodimer from sulfolobus solfataricus | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, DNA/RNA-BINDING PROTEIN ALBA 1, DNA/RNA-BINDING PROTEIN ALBA 2 | | Authors: | Jelinska, C, Conroy, M.J, Craven, C.J, Bullough, P.A, Waltho, J.P, Taylor, G.L, White, M.F. | | Deposit date: | 2005-02-22 | | Release date: | 2005-07-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Obligate Heterodimerization of the Archaeal Alba2 Protein with Alba1 Provides a Mechanism for Control of DNA Packaging.
Structure, 13, 2005
|
|
2BHN
 
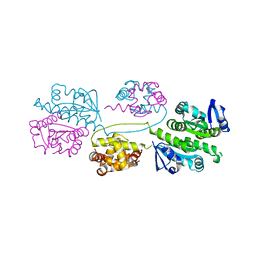 | | XPF from Aeropyrum pernix | | Descriptor: | XPF ENDONUCLEASE | | Authors: | Newman, M, Murray-Rust, J, Lally, J, Rudolf, J, Fadden, A, Knowles, P.P, White, M.F, McDonald, N.Q. | | Deposit date: | 2005-01-14 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of an XPF endonuclease with and without DNA suggests a model for substrate recognition.
EMBO J., 24, 2005
|
|
2BKE
 
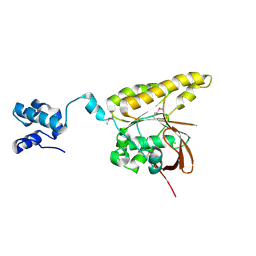 | | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal RadA | | Descriptor: | CHLORIDE ION, DNA REPAIR AND RECOMBINATION PROTEIN RADA | | Authors: | Ariza, A, Richard, D.L, White, M.F, Bond, C.S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal Rada
Nucleic Acids Res., 33, 2005
|
|
2X5F
 
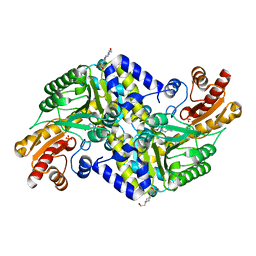 | | Crystal structure of the methicillin-resistant Staphylococcus aureus Sar2028, an aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate dependent aminotransferase | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ASPARTATE_TYROSINE_PHENYLALANINE PYRIDOXAL-5' PHOSPHATE-DEPENDENT AMINOTRANSFERASE, MAGNESIUM ION, ... | | Authors: | Oke, M, Carter, L.G, Johnson, K.A, Liu, H, Mcmahon, S.A, White, M.F, Naismith, J.H. | | Deposit date: | 2010-02-08 | | Release date: | 2010-07-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Scottish Structural Proteomics Facility: Targets, Methods and Outputs.
J.Struct.Funct.Genom., 11, 2010
|
|
1H0X
 
 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
1H0Y
 
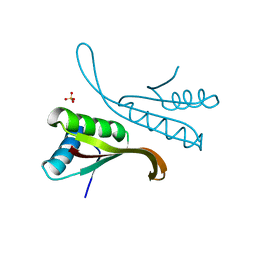 | | Structure of Alba: an archaeal chromatin protein modulated by acetylation | | Descriptor: | DNA BINDING PROTEIN SSO10B, SULFATE ION | | Authors: | Wardleworth, B.N, Russell, R.J.M, Bell, S.D, Taylor, G.L, White, M.F. | | Deposit date: | 2002-07-01 | | Release date: | 2002-09-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Alba: An Archaeal Chromatin Protein Modulated by Acetylation
Embo J., 21, 2002
|
|
