8DYR
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-04 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E17
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P,6M)-6-[1-(2-fluoroethyl)-1H-1,2,3-triazol-4-yl]-4-[5-(methanesulfonyl)-2-methoxyphenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-09 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
8E3W
 
 | | BRD4-D1 in complex with BET inhibitor | | Descriptor: | (4P)-4-[2-(cyclopropylmethoxy)-5-(methanesulfonyl)phenyl]-2-methylisoquinolin-1(2H)-one, 1,2-ETHANEDIOL, Bromodomain-containing protein 4 | | Authors: | Gorman, M.A, Fitzgerald, C.G.D, White, J.M, Parker, M.W. | | Deposit date: | 2022-08-17 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Bromodomain and extraterminal protein-targeted probe enables tumour visualisation in vivo using positron emission tomography.
Chem.Commun.(Camb.), 59, 2023
|
|
4Z4B
 
 | |
5T42
 
 | | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity | | Descriptor: | Envelope glycoprotein | | Authors: | Lee, J, Nyenhuis, D.A, Nelson, E.A, Cafiso, D.S, White, J.M, Tamm, L.K. | | Deposit date: | 2016-08-28 | | Release date: | 2017-08-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4CD6
 
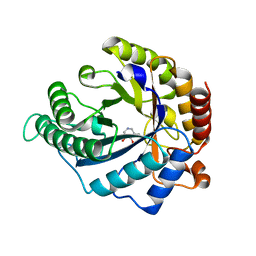 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD7
 
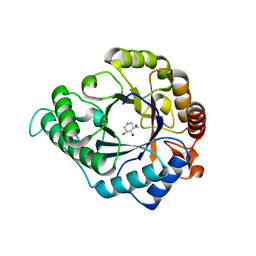 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManIFG and beta-1,4-mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD8
 
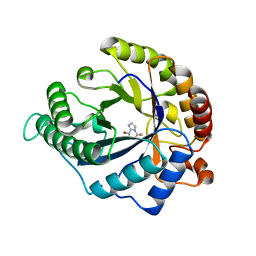 | | The structure of GH113 beta-mannanase AaManA from Alicyclobacillus acidocaldarius in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-BETA-1,4-MANNANASE, beta-D-mannopyranose | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD5
 
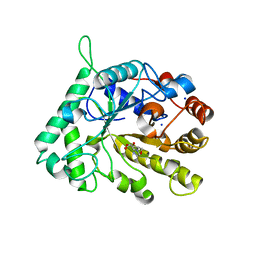 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManMIm | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4CD4
 
 | | The structure of GH26 beta-mannanase CjMan26C from Cellvibrio japonicus in complex with ManIFG | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDO-1,4-BETA MANNANASE, PUTATIVE, ... | | Authors: | Williams, R.J, Iglesias-Fernandez, J, Stepper, J, Jackson, A, Thompson, A.J, Lowe, E.C, White, J.M, Gilbert, H.J, Rovira, C, Davies, G.J, Williams, S.J. | | Deposit date: | 2013-10-30 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Combined Inhibitor Free-Energy Landscape and Structural Analysis Reports on the Mannosidase Conformational Coordinate.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1DIR
 
 | | CRYSTAL STRUCTURE OF A MONOCLINIC FORM OF DIHYDROPTERIDINE REDUCTASE FROM RAT LIVER | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Su, Y, Skinner, M.M, Matthews, D.A, Whitely, J.M, Xuong, N.H. | | Deposit date: | 1994-04-18 | | Release date: | 1994-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a monoclinic form of dihydropteridine reductase from rat liver.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1DHR
 
 | | CRYSTAL STRUCTURE OF RAT LIVER DIHYDROPTERIDINE REDUCTASE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Skinner, M.M, Whiteley, J.M, Matthews, D.A, Xuong, N.H. | | Deposit date: | 1992-03-30 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of rat liver dihydropteridine reductase.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
1HDR
 
 | | THE CRYSTALLOGRAPHIC STRUCTURE OF A HUMAN DIHYDROPTERIDINE REDUCTASE NADH BINARY COMPLEX EXPRESSED IN ESCHERICHIA COLI BY A CDNA CONSTRUCTED FROM ITS RAT HOMOLOGUE | | Descriptor: | DIHYDROPTERIDINE REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Varughese, K.I, Su, Y, Xuong, N.H, Whiteley, J.M. | | Deposit date: | 1993-08-18 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of a human dihydropteridine reductase NADH binary complex expressed in Escherichia coli by a cDNA constructed from its rat homologue.
J.Biol.Chem., 268, 1993
|
|
1SRR
 
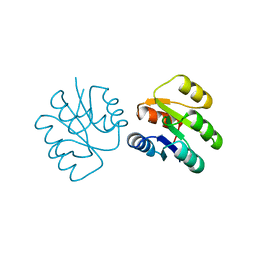 | | CRYSTAL STRUCTURE OF A PHOSPHATASE RESISTANT MUTANT OF SPORULATION RESPONSE REGULATOR SPO0F FROM BACILLUS SUBTILIS | | Descriptor: | CALCIUM ION, SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Whiteley, J.M, Hoch, J.A, Zapf, J, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1996-04-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a phosphatase-resistant mutant of sporulation response regulator Spo0F from Bacillus subtilis.
Structure, 4, 1996
|
|
1NAT
 
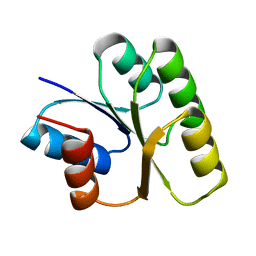 | | CRYSTAL STRUCTURE OF SPOOF FROM BACILLUS SUBTILIS | | Descriptor: | SPORULATION RESPONSE REGULATORY PROTEIN | | Authors: | Madhusudan, Zapf, J, Hoch, J.A, Whiteley, J.M, Xuong, N.H, Varughese, K.I. | | Deposit date: | 1997-09-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis.
Biochemistry, 36, 1997
|
|
1P33
 
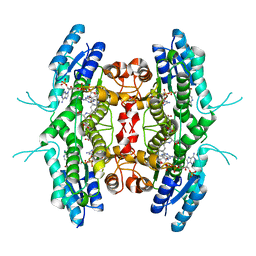 | | Pteridine reductase from Leishmania tarentolae complex with NADPH and MTX | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pteridine reductase 1 | | Authors: | Zhao, H, Bray, T, Ouellette, M, Zhao, M, Ferre, R.A, Matthews, D, Whiteley, J.M, Varughese, K.I. | | Deposit date: | 2003-04-16 | | Release date: | 2003-09-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structure of pteridine reductase (PTR1) from Leishmania tarentolae.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
