2CEO
 
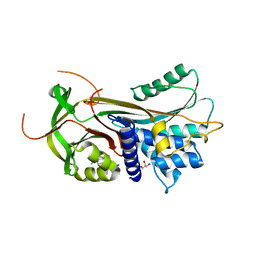 | | thyroxine-binding globulin complex with thyroxine | | Descriptor: | 3,5,3',5'-TETRAIODO-L-THYRONINE, GLYCEROL, THYROXINE-BINDING GLOBULIN | | Authors: | Zhou, A, Wei, Z, Read, R.J, Carrell, R.W. | | Deposit date: | 2006-02-08 | | Release date: | 2006-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Mechanism for the Carriage and Release of Thyroxine in the Blood.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2B1M
 
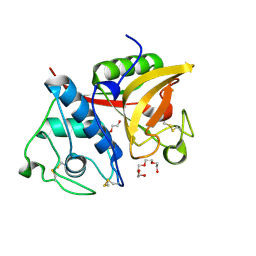 | | Crystal structure of a papain-fold protein without the catalytic cysteine from seeds of Pachyrhizus erosus | | Descriptor: | DI(HYDROXYETHYL)ETHER, SPE31, TETRAETHYLENE GLYCOL, ... | | Authors: | Zhang, M, Wei, Z, Chang, S. | | Deposit date: | 2005-09-16 | | Release date: | 2006-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a papain-fold protein without the catalytic residue: a novel member in the cysteine proteinase family
J.Mol.Biol., 358, 2006
|
|
4JHR
 
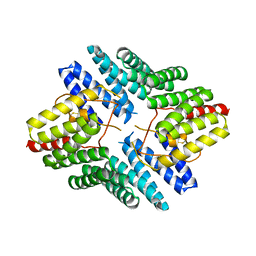 | | An auto-inhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs | | Descriptor: | G-protein-signaling modulator 2 | | Authors: | Pan, Z, Zhu, J, Shang, Y, Wei, Z, Jia, M, Xia, C, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs
Structure, 21, 2013
|
|
7Y8W
 
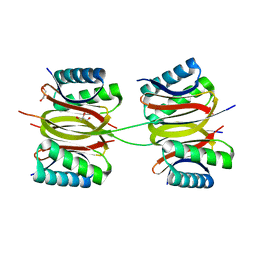 | | Crystal structure of DLC-1/SAO-1 complex | | Descriptor: | Dynein light chain 1, cytoplasmic, GLYCEROL, ... | | Authors: | Yan, H, Zhao, C, Wei, Z, Yu, C. | | Deposit date: | 2022-06-24 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between DLC-1 and SAO-1 facilitates CED-4 translocation during apoptosis in the Caenorhabditis elegans germline.
Cell Death Discov, 8, 2022
|
|
7YV9
 
 | |
6D26
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with fevipiprant | | Descriptor: | OLEIC ACID, Prostaglandin D2 receptor 2, Endolysin chimera, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6C1R
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MALONATE ION, OLEIC ACID, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D27
 
 | | Crystal structure of the prostaglandin D2 receptor CRTH2 with CAY10471 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, OLEIC ACID, ... | | Authors: | Wang, L, Yao, D, Deepak, K, Liu, H, Gong, W, Fan, H, Wei, Z, Zhang, C. | | Deposit date: | 2018-04-13 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.738 Å) | | Cite: | Structures of the Human PGD2Receptor CRTH2 Reveal Novel Mechanisms for Ligand Recognition.
Mol. Cell, 72, 2018
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4AFX
 
 | | Crystal structure of the reactive loop cleaved ZPI in I2 space group | | Descriptor: | CALCIUM ION, PHOSPHATE ION, PROTEIN Z DEPENDENT PROTEASE INHIBITOR, ... | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-01-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
4AJU
 
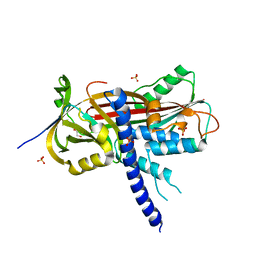 | | Crystal structure of the reactive loop cleaved ZPI in P41 space group | | Descriptor: | 1,2-ETHANEDIOL, PROTEIN Z-DEPENDENT PROTEASE INHIBITOR, SULFATE ION | | Authors: | Zhou, A, Yan, Y, Wei, Z. | | Deposit date: | 2012-02-19 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis for Catalytic Activation of Protein Z-Dependent Protease Inhibitor (Zpi) by Protein Z.
Blood, 120, 2012
|
|
8W41
 
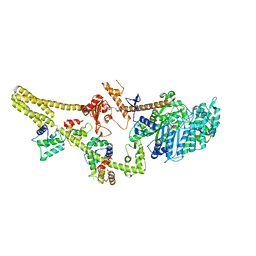 | | Cryo-EM structure of Myosin VI in the autoinhibited state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Calmodulin-1, ... | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2023-08-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Autoinhibition and activation of myosin VI revealed by its cryo-EM structure.
Nat Commun, 15, 2024
|
|
8WHI
 
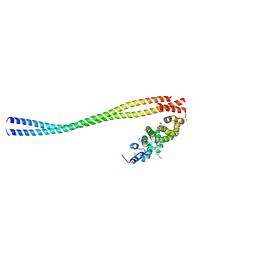 | |
8WHK
 
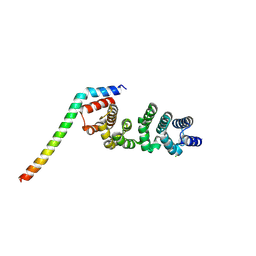 | | Crystal structure of CLASP2 in complex with LL5beta | | Descriptor: | CHLORIDE ION, CLIP-associating protein 2, Pleckstrin homology-like domain family B member 2 | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHJ
 
 | | Crystal structure of CLASP2 TOG4 fused with LL5beta | | Descriptor: | CHLORIDE ION, CLIP-associating protein 2,Pleckstrin homology-like domain family B member 2, DI(HYDROXYETHYL)ETHER | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHL
 
 | | Crystal structure of CLASP2 in complex with CENP-E | | Descriptor: | ACETATE ION, CLIP-associating protein 2, Centromere-associated protein E, ... | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
8WHH
 
 | |
8WHM
 
 | | Crystal structure of the ELKS2/LL5beta complex | | Descriptor: | ERC protein 2, Pleckstrin homology-like domain family B member 2 | | Authors: | Jia, X, Wei, Z. | | Deposit date: | 2023-09-23 | | Release date: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CLASP-mediated competitive binding in protein condensates directs microtubule growth.
Nat Commun, 15, 2024
|
|
3TFM
 
 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
6F4V
 
 | |
6F4U
 
 | |
6KU0
 
 | | Crystal structure of MyoVa-GTD in complex with MICAL1-GTBM | | Descriptor: | 1,2-ETHANEDIOL, Peptide from [F-actin]-monooxygenase MICAL1, Unconventional myosin-Va | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.
Sci Adv, 6, 2020
|
|
3G5B
 
 | | The structure of UNC5b cytoplasmic domain | | Descriptor: | Netrin receptor UNC5B, PHOSPHATE ION | | Authors: | Wang, R, Wei, Z, Zhang, M. | | Deposit date: | 2009-02-04 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Autoinhibition of UNC5b revealed by the cytoplasmic domain structure of the receptor
Mol.Cell, 33, 2009
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPY
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
