2FQF
 
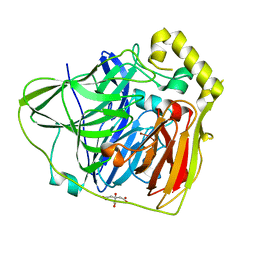 | | Crystal Structures of E. coli Laccase CueO under different copper binding situations | | Descriptor: | Blue copper oxidase cueO, CITRIC ACID, COPPER (II) ION, ... | | Authors: | Li, X, Wei, Z, Zhang, M, Teng, M, Gong, W. | | Deposit date: | 2006-01-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli laccase CueO at different copper concentrations.
Biochem.Biophys.Res.Commun., 354, 2007
|
|
4JHR
 
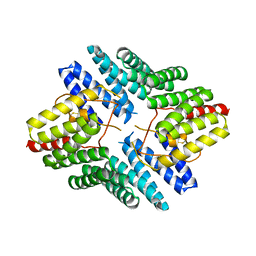 | | An auto-inhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs | | Descriptor: | G-protein-signaling modulator 2 | | Authors: | Pan, Z, Zhu, J, Shang, Y, Wei, Z, Jia, M, Xia, C, Wen, W, Wang, W, Zhang, M. | | Deposit date: | 2013-03-05 | | Release date: | 2013-06-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An autoinhibited conformation of LGN reveals a distinct interaction mode between GoLoco motifs and TPR motifs
Structure, 21, 2013
|
|
6A68
 
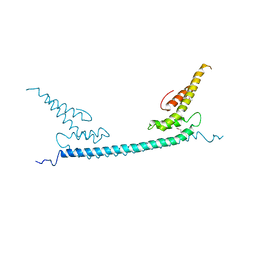 | | the crystal structure of rat calcium-dependent activator protein for secretion (CAPS) DAMH domain | | Descriptor: | Calcium-dependent secretion activator 1, POTASSIUM ION | | Authors: | Zhou, H, Wei, Z.Q, Yao, D.Q, Zhang, R.G, Ma, C. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structural and Functional Analysis of the CAPS SNARE-Binding Domain Required for SNARE Complex Formation and Exocytosis.
Cell Rep, 26, 2019
|
|
4MPL
 
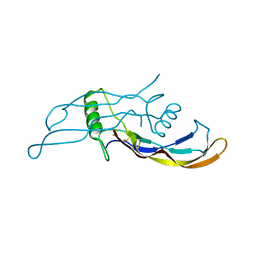 | | Crystal structure of BMP9 at 1.90 Angstrom | | Descriptor: | Growth/differentiation factor 2 | | Authors: | Li, W, Morrell, N.W, Wei, Z. | | Deposit date: | 2013-09-13 | | Release date: | 2014-09-17 | | Last modified: | 2021-06-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Regulation of Bone Morphogenetic Protein 9 (BMP9) by Redox-dependent Proteolysis.
J.Biol.Chem., 289, 2014
|
|
8HLO
 
 | | Crystal structure of ASAP1-SH3 and MICAL1-PRM complex | | Descriptor: | Arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 1, Proline rich motif from MICAL1, ... | | Authors: | Niu, F, Wei, Z, Yu, C. | | Deposit date: | 2022-11-30 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.168 Å) | | Cite: | Crystal Structure of the SH3 Domain of ASAP1 in Complex with the Proline Rich Motif (PRM) of MICAL1 Reveals a Unique SH3/PRM Interaction Mode.
Int J Mol Sci, 24, 2023
|
|
2I6K
 
 | | Crystal structure of human type I IPP isomerase complexed with a substrate analog | | Descriptor: | ACETIC ACID, AMINOETHANOLPYROPHOSPHATE, Isopentenyl-diphosphate delta-isomerase 1, ... | | Authors: | Zhang, C, Wei, Z, Gong, W. | | Deposit date: | 2006-08-29 | | Release date: | 2007-06-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of human IPP isomerase: new insights into the catalytic mechanism
J.Mol.Biol., 366, 2007
|
|
2LE8
 
 | | The protein complex for DNA replication | | Descriptor: | DNA replication factor Cdt1, DNA replication licensing factor MCM6 | | Authors: | Liu, C, Wei, Z, Zhu, G. | | Deposit date: | 2011-06-14 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural insights into the Cdt1-mediated MCM2-7 chromatin loading
Nucleic Acids Res., 40, 2012
|
|
2L7T
 
 | |
2LW7
 
 | | NMR solution structure of human HisRS splice variant | | Descriptor: | Histidine--tRNA ligase, cytoplasmic | | Authors: | Ye, F, Wei, Z, Wu, J, Schimmel, P, Zhang, M. | | Deposit date: | 2012-07-24 | | Release date: | 2013-09-04 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of human HisRS splice variant
To be Published
|
|
5XQ1
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-3 | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPY
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | ACETATE ION, Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XPZ
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5XQ0
 
 | | Structural basis of kindlin-mediated integrin recognition and activation | | Descriptor: | Fermitin family homolog 2,Integrin beta-1, GLYCEROL | | Authors: | Li, H, Yang, H, Sun, K, Zhang, Z, Yu, C, Wei, Z. | | Deposit date: | 2017-06-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of kindlin-mediated integrin recognition and activation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8IJ9
 
 | | Crystal structure of the ELKS1/Rab6B complex | | Descriptor: | ELKS/Rab6-interacting/CAST family member 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jin, G, Wei, Z. | | Deposit date: | 2023-02-26 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of ELKS/Rab6B interaction and its role in vesicle capturing enhanced by liquid-liquid phase separation.
J.Biol.Chem., 299, 2023
|
|
8IW5
 
 | | Crystal structure of liprin-beta H2H3 dimer | | Descriptor: | CALCIUM ION, Liprin-beta-1 | | Authors: | Zhang, J, Chen, S, Wei, Z. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
8IW0
 
 | | Crystal structure of the KANK1/liprin-beta1 complex | | Descriptor: | Liprin-beta-1,KN motif and ankyrin repeat domain-containing protein 1 | | Authors: | Zhang, J, Chen, S, Wei, Z, Yu, C. | | Deposit date: | 2023-03-29 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | KANK1 shapes focal adhesions by orchestrating protein binding, mechanical force sensing, and phase separation.
Cell Rep, 42, 2023
|
|
3TFM
 
 | | Myosin X PH1N-PH2-PH1C tandem | | Descriptor: | Myosin X, PHOSPHATE ION | | Authors: | Yu, J, Lu, Q, Yan, J, Wei, Z, Zhang, M. | | Deposit date: | 2011-08-16 | | Release date: | 2011-12-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural basis of the myosin X PH1N-PH2-PH1C tandem as a specific and acute cellular PI(3,4,5)P3 sensor
MOLECULAR BIOLOGY OF THE CELL, 22, 2011
|
|
2KIA
 
 | | Solution structure of Myosin VI C-terminal cargo-binding domain | | Descriptor: | Myosin-VI | | Authors: | Feng, W, Yu, C, Wei, Z, Miyanoiri, Y, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
3NDP
 
 | | Crystal structure of human AK4(L171P) | | Descriptor: | Adenylate kinase isoenzyme 4, SULFATE ION | | Authors: | Liu, R, Wang, Y, Wei, Z, Gong, W. | | Deposit date: | 2010-06-07 | | Release date: | 2010-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human adenylate kinase 4 (L171P) suggests the role of hinge region in protein domain motion
Biochem.Biophys.Res.Commun., 379, 2009
|
|
6C1R
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist avacopan | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, MALONATE ION, OLEIC ACID, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6C1Q
 
 | | Crystal structure of human C5a receptor in complex with an orthosteric antagonist PMX53 and an allosteric antagonist NDT9513727 | | Descriptor: | 1-(1,3-benzodioxol-5-yl)-~{N}-(1,3-benzodioxol-5-ylmethyl)-~{N}-[(3-butyl-2,5-diphenyl-imidazol-4-yl)methyl]methanamine, PMX53, Soluble cytochrome b562, ... | | Authors: | Liu, H, Wang, L, Wei, Z, Zhang, C. | | Deposit date: | 2018-01-05 | | Release date: | 2018-05-30 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Orthosteric and allosteric action of the C5a receptor antagonists.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6LR3
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2020-01-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6LKV
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor, SULFATE ION | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6LKW
 
 | | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum | | Descriptor: | CHLORIDE ION, Macrophage migration inhibitory factor | | Authors: | Su, Z.M, Tian, X.Y, Li, H.J, Wei, Z.M, Chen, L.F, Ren, H.X, Peng, W.F, Tang, C.T. | | Deposit date: | 2019-12-20 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and functional insights into macrophage migration inhibitory factor from Oncomelania hupensis, the intermediate host of Schistosoma japonicum.
Biochem.J., 477, 2020
|
|
6KU0
 
 | | Crystal structure of MyoVa-GTD in complex with MICAL1-GTBM | | Descriptor: | 1,2-ETHANEDIOL, Peptide from [F-actin]-monooxygenase MICAL1, Unconventional myosin-Va | | Authors: | Niu, F, Wei, Z. | | Deposit date: | 2019-08-29 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | F-actin disassembly factor MICAL1 binding to Myosin Va mediates cargo unloading during cytokinesis.
Sci Adv, 6, 2020
|
|
