8V83
 
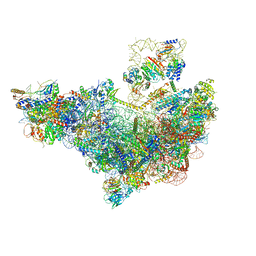 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.53 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
6B5T
 
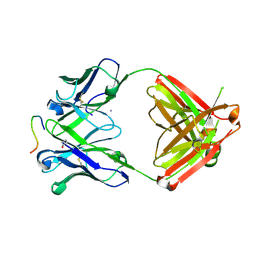 | | Structure of PfCSP peptide 29 with human antibody CIS42 | | Descriptor: | AMMONIUM ION, CIS42 Fab Heavy chain, CIS42 Fab Light chain, ... | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.222 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6B5S
 
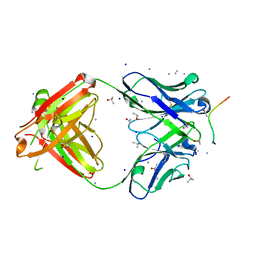 | | Structure of PfCSP peptide 25 with human antibody CIS42 | | Descriptor: | AMMONIUM ION, CIS42 Fab Heavy chain, CIS42 Fab Light chain, ... | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2019-01-09 | | Method: | X-RAY DIFFRACTION (1.983 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6C5V
 
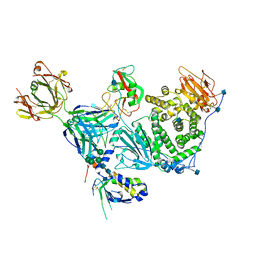 | | An anti-gH/gL antibody that neutralizes dual-tropic infection defines a site of vulnerability on Epstein-Barr virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab AMMO1 heavy chain, Antibody Fab AMMO1 light chain, ... | | Authors: | Snijder, J, Ortego, M.S, Weidle, C, Stuart, A.B, Gray, M.A, McElrath, M.J, Pancera, M, Veesler, D, McGuire, A.T, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2018-01-16 | | Release date: | 2018-05-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
6B5R
 
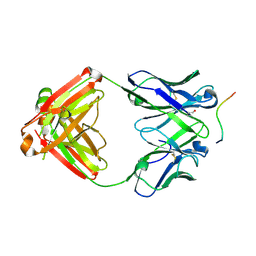 | | Structure of PfCSP peptide 21 with human antibody CIS42 | | Descriptor: | CIS42 Fab Heavy chain, CIS42 Fab Light chain, PfCSP peptide 21: ASN-PRO-ASP-PRO-ASN-ALA-ASN-PRO-ASN-VAL-ASP-PRO-ASN | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (1.775 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6B5P
 
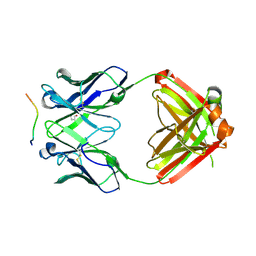 | | Structure of PfCSP peptide 20 with human antibody CIS42 | | Descriptor: | CIS42 Fab Heavy chain, CIS42 Fab Light chain, pfCSP peptide 20: ASN-PRO-ASP-PRO-ASN-ALA-ASN-PRO-ASN-VAL-ASP | | Authors: | Pancera, M, Weidle, C. | | Deposit date: | 2017-09-29 | | Release date: | 2018-03-21 | | Last modified: | 2018-04-25 | | Method: | X-RAY DIFFRACTION (2.297 Å) | | Cite: | A human monoclonal antibody prevents malaria infection by targeting a new site of vulnerability on the parasite.
Nat. Med., 24, 2018
|
|
6BLA
 
 | | Structure of AMM01 Fab, an anti EBV gH/gL neutralizing antibody | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMM01 Fab Heavy chain, ... | | Authors: | Pancera, M, Weidle, C, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2017-11-09 | | Release date: | 2018-04-25 | | Last modified: | 2019-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An Antibody Targeting the Fusion Machinery Neutralizes Dual-Tropic Infection and Defines a Site of Vulnerability on Epstein-Barr Virus.
Immunity, 48, 2018
|
|
1PLG
 
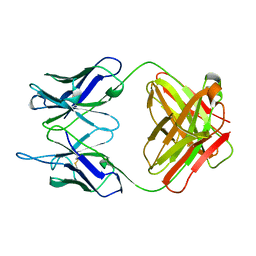 | | EVIDENCE FOR THE EXTENDED HELICAL NATURE OF POLYSACCHARIDE EPITOPES. THE 2.8 ANGSTROMS RESOLUTION STRUCTURE AND THERMODYNAMICS OF LIGAND BINDING OF AN ANTIGEN BINDING FRAGMENT SPECIFIC FOR ALPHA-(2->8)-POLYSIALIC ACID | | Descriptor: | IGG2A=KAPPA= | | Authors: | Evans, S.V, Sigurskjold, B.W, Jennings, H.J, Brisson, J.-R, Tse, W.C, To, R, Altman, E, Frosch, M, Weisgerber, C, Kratzin, H, Klebert, S, Vaesen, M, Bitter-Suermann, D, Rose, D.R, Young, N.M, Bundle, D.R. | | Deposit date: | 1995-04-24 | | Release date: | 1996-04-03 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evidence for the extended helical nature of polysaccharide epitopes. The 2.8 A resolution structure and thermodynamics of ligand binding of an antigen binding fragment specific for alpha-(2-->8)-polysialic acid.
Biochemistry, 34, 1995
|
|
4A4V
 
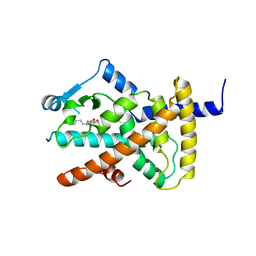 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin 2 | | Descriptor: | AMORFRUTIN 2, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
4A4W
 
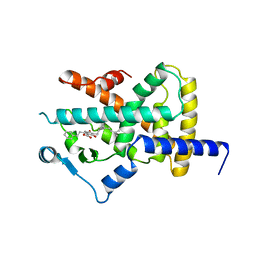 | | Ligand binding domain of human PPAR gamma in complex with amorfrutin B | | Descriptor: | AMORFRUTIN B, PEROXISOME PROLIFERATOR-ACTIVATED RECEPTOR GAMMA | | Authors: | de Groot, J.C, Weidner, C, Krausze, J, Kawamoto, K, Schroeder, F.C, Sauer, S, Buessow, K. | | Deposit date: | 2011-10-20 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Characterization of Amorfrutins Bound to the Peroxisome Proliferator-Activated Receptor Gamma.
J.Med.Chem., 56, 2013
|
|
7UQB
 
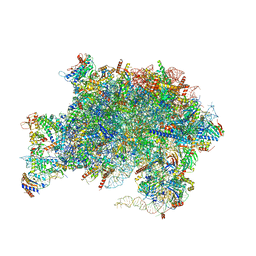 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1-D52A strain with AlF4 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UOO
 
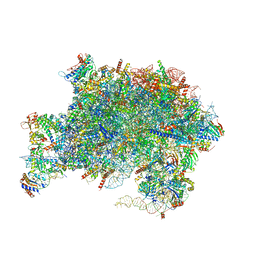 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7UQZ
 
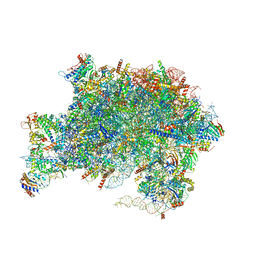 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1 D52A strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7V08
 
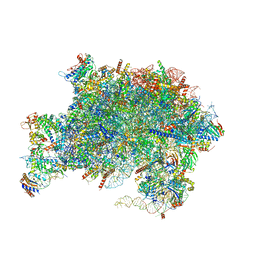 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a Spb1 D52A suppressor 3 strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-05-10 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
7U63
 
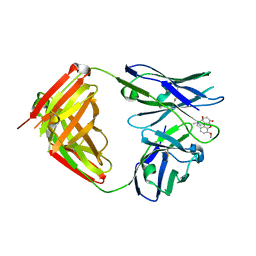 | | Crystal Structure Anti-Oxycodone Antibody HY2-A12 Fab Complexed with Oxycodone | | Descriptor: | 14-hydroxy-3-methoxy-17-methyl-5beta-4,5-epoxymorphinan-6-one, HY2-A12 Fab Heavy Chain, HY2-A12 Fab Light Chain | | Authors: | Rodarte, J.V, Pancera, M.P, Weidle, C, Rupert, P.B, Strong, R.K. | | Deposit date: | 2022-03-03 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding.
Structure, 31, 2023
|
|
8V3B
 
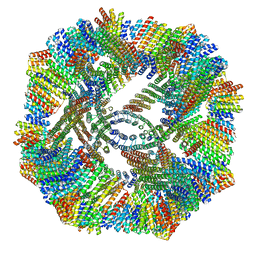 | |
1FX8
 
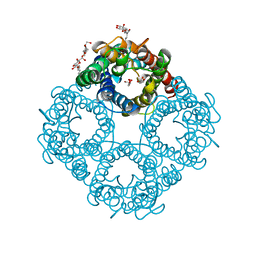 | | CRYSTAL STRUCTURE OF THE E. COLI GLYCEROL FACILITATOR (GLPF) WITH SUBSTRATE GLYCEROL | | Descriptor: | GLYCEROL, GLYCEROL UPTAKE FACILITATOR PROTEIN, octyl beta-D-glucopyranoside | | Authors: | Fu, D, Libson, A, Miercke, L.J.W, Weitzman, C, Nollert, P, Stroud, R.M. | | Deposit date: | 2000-09-25 | | Release date: | 2000-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a glycerol-conducting channel and the basis for its selectivity.
Science, 290, 2000
|
|
7UUI
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing post-rotation state from a SPB1 D52A strain | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleolar GTP-binding protein 2, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-28 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
8V85
 
 | |
5VET
 
 | | PHOSPHOLIPASE A2, RE-REFINEMENT OF THE PDB STRUCTURE 1JQ8 WITHOUT THE PUTATIVE COMPLEXED OLIGOPEPTIDE | | Descriptor: | Phospholipase A2 VRV-PL-VIIIa | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2018-06-20 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
6HAQ
 
 | | AFGH61B WILD-TYPE COPPER LOADED | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (II) ION, ... | | Authors: | Lo Leggio, L, Poulsen, J.C.N, Weihe, C.D. | | Deposit date: | 2018-08-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structure of a lytic polysaccharide monooxygenase from Aspergillus fumigatus and an engineered thermostable variant.
Carbohydr. Res., 469, 2018
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEH
 
 | | Re-refinement OF THE PDB STRUCTURE 1yiz of Aedes aegypti kynurenine aminotransferase | | Descriptor: | BROMIDE ION, GLYCEROL, Kynurenine aminotransferase | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-04 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
