1ELJ
 
 | | THE CRYSTAL STRUCTURE OF LIGANDED MALTODEXTRIN-BINDING PROTEIN FROM PYROCOCCUS FURIOSUS | | Descriptor: | MALTODEXTRIN-BINDING PROTEIN, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Evdokimov, A.G, Anderson, E.D, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-03-13 | | Release date: | 2001-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for oligosaccharide recognition by Pyrococcus furiosus maltodextrin-binding protein.
J.Mol.Biol., 305, 2001
|
|
6DJJ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DJG
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ503 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxy-8-sulfoquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6DIH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with Sigma Aldrich compound PH004941 | | Descriptor: | 1,2-ETHANEDIOL, 4-hydroxybenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
3MMG
 
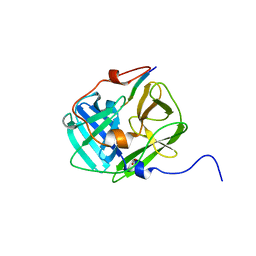 | | Crystal structure of tobacco vein mottling virus protease | | Descriptor: | FORMIC ACID, Nuclear inclusion protein A, Nuclear inclusion protein B fragment | | Authors: | Ping, S, Austin, B.P, Tozser, J, Waugh, D.S. | | Deposit date: | 2010-04-19 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants of tobacco vein mottling virus protease substrate specificity.
Protein Sci., 19, 2010
|
|
6XBC
 
 | |
6XBB
 
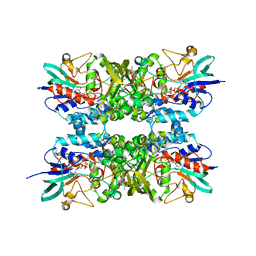 | |
1RFA
 
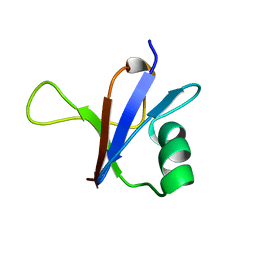 | | NMR SOLUTION STRUCTURE OF THE RAS-BINDING DOMAIN OF C-RAF-1 | | Descriptor: | RAF1 | | Authors: | Emerson, S.D, Madison, V.S, Palermo, R.E, Waugh, D.S, Scheffler, J.E, Tsao, K.-L, Kiefer, S.E, Liu, S.P, Fry, D.C. | | Deposit date: | 1995-04-26 | | Release date: | 1996-06-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ras-binding domain of c-Raf-1 and identification of its Ras interaction surface.
Biochemistry, 34, 1995
|
|
4A0E
 
 | |
4AZ1
 
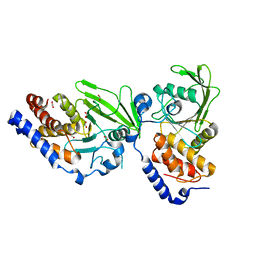 | | Crystal structure of the Trypanosoma cruzi protein tyrosine phosphatase TcPTP1, a potential therapeutic target for Chagas' disease | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, TYROSINE SPECIFIC PROTEIN PHOSPHATASE | | Authors: | Lountos, G.T, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2012-06-22 | | Release date: | 2012-11-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.181 Å) | | Cite: | Structure of the Trypanosoma Cruzi Protein Tyrosine Phosphatase Tcptp1, a Potential Therapeutic Target for Chagas' Disease.
Mol.Biochem.Parasitol., 187, 2012
|
|
2AOB
 
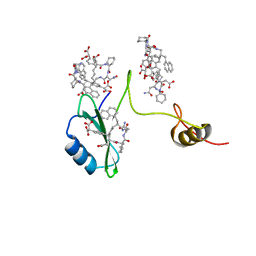 | | Crystal structures of a high-affinity macrocyclic peptide mimetic in complex with the Grb2 SH2 domain | | Descriptor: | 2-(4-((9S,10S,14S,Z)-18-(2-AMINO-2-OXOETHYL)-9-(CARBOXYMETHYL)-14-(NAPHTHALEN-1-YLMETHYL)-8,17,20-TRIOXO-7,16,19-TRIAZASPIRO[5.14]ICOS-11-EN-10-YL)PHENYL)MALONIC ACID, Growth factor receptor-bound protein 2 | | Authors: | Phan, J, Shi, Z.D, Burke, T.R, Waugh, D.S. | | Deposit date: | 2005-08-12 | | Release date: | 2005-10-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of a High-affinity Macrocyclic Peptide Mimetic in Complex with the Grb2 SH2 Domain.
J.Mol.Biol., 353, 2005
|
|
2EZ6
 
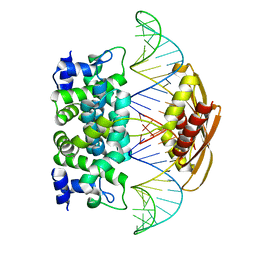 | | Crystal structure of Aquifex aeolicus RNase III (D44N) complexed with product of double-stranded RNA processing | | Descriptor: | 28-MER, MAGNESIUM ION, Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-11-10 | | Release date: | 2006-02-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insight into the Mechanism of Double-Stranded RNA Processing by Ribonuclease III.
Cell(Cambridge,Mass.), 124, 2006
|
|
6W4R
 
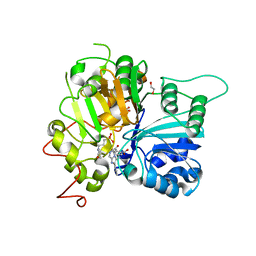 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ633p | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyrazin-3-yl]amino}benzene-1,2-dicarboxylic acid, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
6W7L
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ632p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyrazin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
6W7K
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ634p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyridin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
6W7J
 
 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ635p | | Descriptor: | 1,2-ETHANEDIOL, 4-{[2-(2-hydroxyphenyl)imidazo[1,2-a]pyridin-3-yl]amino}benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Tdp1 catalytic domain
To Be Published
|
|
2YIQ
 
 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1322 | | Descriptor: | (E)-5-(1-(2-CARBAMIMIDOYLHYDRAZONO)ETHYL)-N-(1H-INDOL-6-YL)-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
2YIR
 
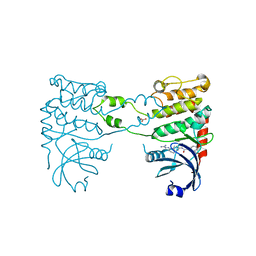 | | Structural analysis of checkpoint kinase 2 in complex with inhibitor PV1352 | | Descriptor: | (E)-N-(5-(2-CARBAMIMIDOYLHYDRAZONO)-5,6,7,8-TETRAHYDRONAPHTHALEN-2-YL)-7-NITRO-1H-INDOLE-2-CARBOXAMIDE, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
2YIT
 
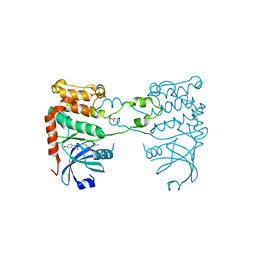 | | Structural analysis of checkpoint kinase 2 in complex with PV1162, a novel inhibitor | | Descriptor: | N-{4-[(1E)-N-carbamimidoylbutanehydrazonoyl]phenyl}-5-methoxy-1H-indole-2-carboxamide, NITRATE ION, SERINE/THREONINE-PROTEIN KINASE CHK2 | | Authors: | Lountos, G.T, Jobson, A.G, Tropea, J.E, Self, C, Zhang, G, Pommier, Y, Shoemaker, R.H, Waugh, D.S. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-Ray Structures of Checkpoint Kinase 2 in Complex with Inhibitors that Target its Gatekeeper-Dependent Hydrophobic Pocket.
FEBS Lett., 585, 2011
|
|
4M2Z
 
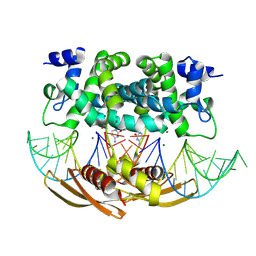 | | Crystal structure of RNASE III complexed with double-stranded RNA and CMP (TYPE II CLEAVAGE) | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, MAGNESIUM ION, RNA10, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
4M30
 
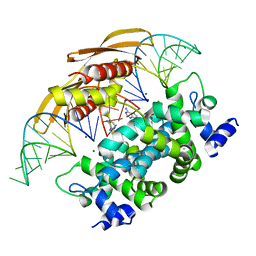 | | Crystal structure of RNASE III complexed with double-stranded RNA AND AMP (TYPE II CLEAVAGE) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Gan, J, Liang, Y.-H, Shaw, G.X, Tropea, J.E, Waugh, D.S, Ji, X. | | Deposit date: | 2013-08-05 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | RNase III: Genetics and Function; Structure and Mechanism.
Annu. Rev. Genet., 47, 2013
|
|
1G9U
 
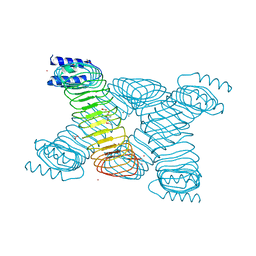 | | CRYSTAL STRUCTURE OF YOPM-LEUCINE RICH EFFECTOR PROTEIN FROM YERSINIA PESTIS | | Descriptor: | ACETATE ION, CALCIUM ION, MERCURY (II) ION, ... | | Authors: | Evdokimov, A.G, Anderson, D.E, Routzahn, K.M, Waugh, D.S. | | Deposit date: | 2000-11-28 | | Release date: | 2001-10-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit.
J.Mol.Biol., 312, 2001
|
|
1HY5
 
 | |
1HUF
 
 | |
4Y2E
 
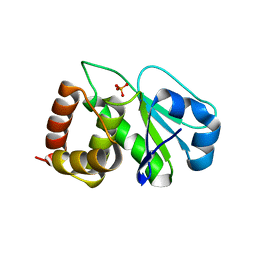 | | Crystal structure of the catalytic domain of human dual-specificity phosphatase 7 (C232S) | | Descriptor: | Dual specificity protein phosphatase 7, PHOSPHATE ION | | Authors: | Lountos, G.T, Austin, B.P, Tropea, J.E, Waugh, D.S. | | Deposit date: | 2015-02-09 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure of human dual-specificity phosphatase 7, a potential cancer drug target.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
