8PA0
 
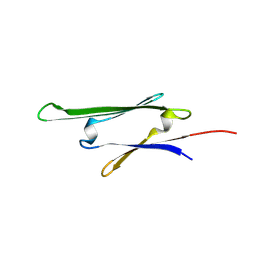 | | cvHsp (HspB7) C131S alpha-crystallin domain - filamin C (FLNC) domain 24 complex | | Descriptor: | Filamin-C, Heat shock protein beta-7 | | Authors: | Wang, Z, Benesch, J.L.P, Allison, T.M, Song, H, McDonough, M.A, Brem, J, Rabe, P. | | Deposit date: | 2023-06-06 | | Release date: | 2024-10-16 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Filamin C dimerisation is regulated by HSPB7.
Nat Commun, 16, 2025
|
|
3FMZ
 
 | |
9IUZ
 
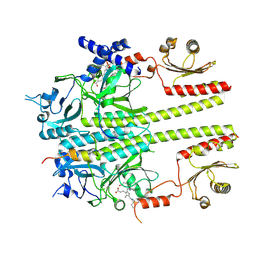 | | Constitutively active mutant(Y276H) of Arabidopsis phytochrome B(phyB) in complex with phytochrome-interacting factor 6(PIF6) | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome B, Phytochrome-interacting factor 6 | | Authors: | Wang, Z, Wang, W, Zhao, D, Song, Y, Xu, B, Zhao, J, Wang, J. | | Deposit date: | 2024-07-22 | | Release date: | 2024-10-02 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Light-induced remodeling of phytochrome B enables signal transduction by phytochrome-interacting factor.
Cell, 187, 2024
|
|
3K8S
 
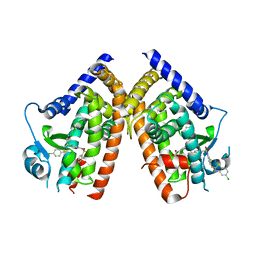 | | Crystal Structure of PPARg in complex with T2384 | | Descriptor: | 2-chloro-N-{3-chloro-4-[(5-chloro-1,3-benzothiazol-2-yl)sulfanyl]phenyl}-4-(trifluoromethyl)benzenesulfonamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Wang, Z. | | Deposit date: | 2009-10-14 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | T2384, a novel antidiabetic agent with unique peroxisome proliferator-activated receptor gamma binding properties
J.Biol.Chem., 283, 2008
|
|
1PBP
 
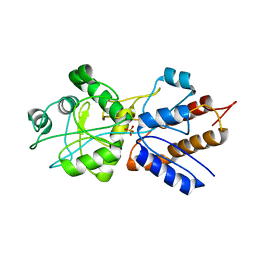 | | FINE TUNING OF THE SPECIFICITY OF THE PERIPLASMIC PHOSPHATE TRANSPORT RECEPTOR: SITE-DIRECTED MUTAGENESIS, LIGAND BINDING, AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Wang, Z, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1994-07-20 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine tuning the specificity of the periplasmic phosphate transport receptor. Site-directed mutagenesis, ligand binding, and crystallographic studies.
J.Biol.Chem., 269, 1994
|
|
4NA4
 
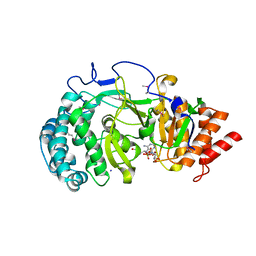 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADP-HPD | | Descriptor: | 5'-O-[(S)-{[(S)-{[(2R,3R,4S)-3,4-DIHYDROXYPYRROLIDIN-2-YL]METHOXY}(HYDROXY)PHOSPHORYL]OXY}(HYDROXY)PHOSPHORYL]ADENOSINE, IODIDE ION, Poly(ADP-ribose) glycohydrolase | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4NA0
 
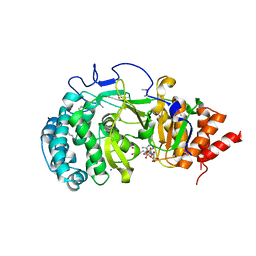 | | Crystal structure of mouse poly(ADP-ribose) glycohydrolase (PARG) catalytic domain with ADPRibose | | Descriptor: | IODIDE ION, Poly(ADP-ribose) glycohydrolase, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Wang, Z, Cheng, Z, Xu, W. | | Deposit date: | 2013-10-21 | | Release date: | 2014-01-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and biochemical analysis of the mouse poly(ADP-ribose) glycohydrolase.
Plos One, 9, 2014
|
|
4N9Y
 
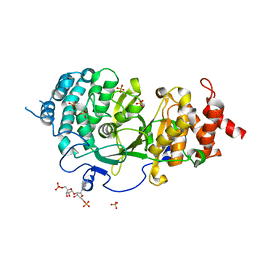 | |
4NA5
 
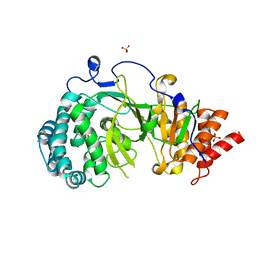 | |
3FRJ
 
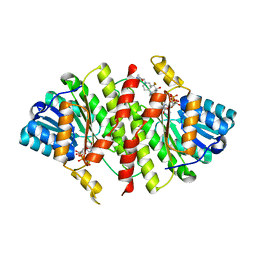 | | Crystal Structure of 11b-Hydroxysteroid Dehydrogenase-1 (11b-HSD1) in Complex with Piperidyl Benzamide Inhibitor | | Descriptor: | Corticosteroid 11-beta-Dehydrogenase, Isozyme 1, N-{1-[(1-carbamoylcyclopropyl)methyl]piperidin-4-yl}-N-cyclopropyl-4-[(1S)-2,2,2-trifluoro-1-hydroxy-1-methylethyl]benzamide, ... | | Authors: | Wang, Z, Sudom, A, Walker, N.P. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and optimization of piperidyl benzamide derivatives as a novel class of 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
9IJN
 
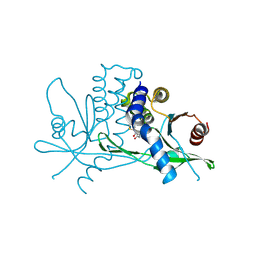 | | STING LBD domain with an agonist DW18343 | | Descriptor: | 4-[4-(7-fluoranyl-1H-indol-3-yl)thiophen-2-yl]-4-oxidanylidene-butanoic acid, Stimulator of interferon genes protein | | Authors: | Wang, Z. | | Deposit date: | 2024-06-25 | | Release date: | 2025-01-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a non-nucleotide stimulator of interferon genes (STING) agonist with systemic antitumor effect.
MedComm (2020), 6, 2025
|
|
5XFO
 
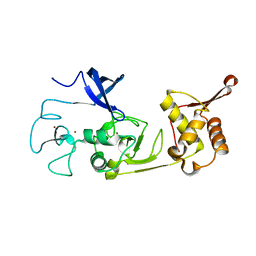 | | Structure of the N-terminal domains of PHF1 | | Descriptor: | PHD finger protein 1, ZINC ION | | Authors: | Wang, Z, Li, H. | | Deposit date: | 2017-04-11 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Polycomb-like proteins link the PRC2 complex to CpG islands
Nature, 549, 2017
|
|
4RF7
 
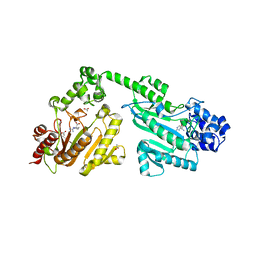 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with substrate L-arginine | | Descriptor: | ACETATE ION, ARGININE, Arginine kinase | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RF8
 
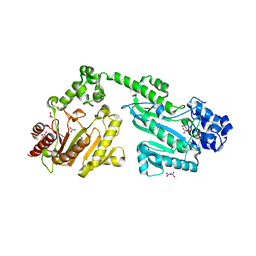 | | Crystal structure of double-domain arginine kinase from Anthopleura japonicas in complex with ADP | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Arginine kinase, ... | | Authors: | Wang, Z, Qiao, Z, Ye, S, Zhang, R. | | Deposit date: | 2014-09-25 | | Release date: | 2015-04-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of a double-domain phosphagen kinase reveals an asymmetric arrangement of the tandem domains.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1IXI
 
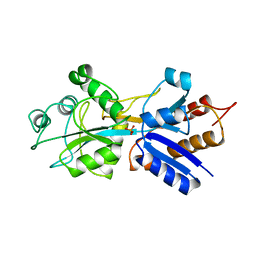 | |
1IXG
 
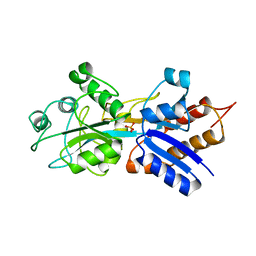 | |
1IXH
 
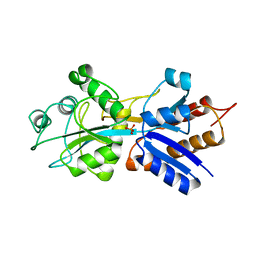 | |
5JJA
 
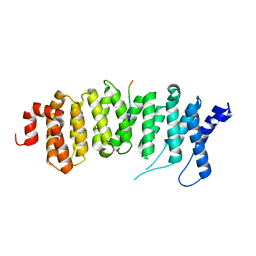 | | Crystal structure of a PP2A B56gamma/BubR1 complex | | Descriptor: | Mitotic checkpoint serine/threonine-protein kinase BUB1 beta, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2016-04-22 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a PP2A B56-BubR1 complex and its implications for PP2A substrate recruitment and localization.
Protein Cell, 7, 2016
|
|
7Q6L
 
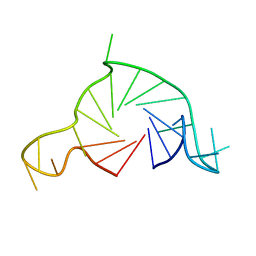 | |
7Q48
 
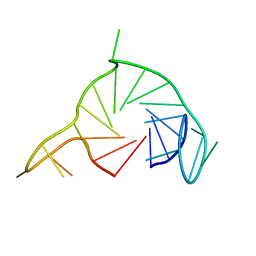 | |
7QA2
 
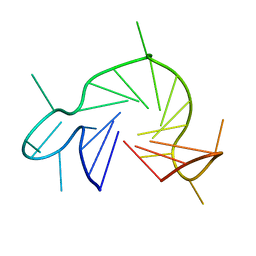 | |
3GL6
 
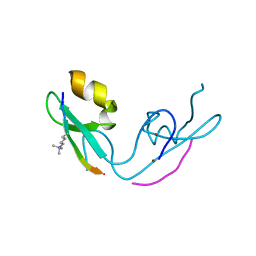 | |
7QIO
 
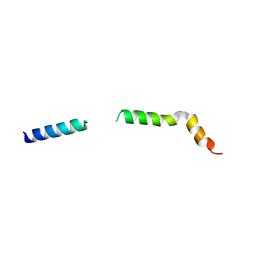 | | Homology model of myosin neck domain in skeletal sarcomere | | Descriptor: | Myosin light chain 1/3, skeletal muscle isoform, Myosin regulatory light chain 2, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
7QIN
 
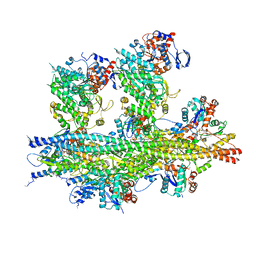 | | In situ structure of actomyosin complex in skeletal sarcomere | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-02-16 | | Last modified: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
7QIM
 
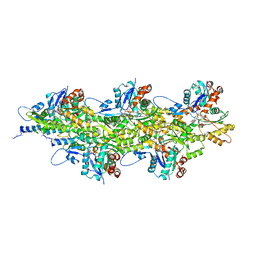 | | In situ structure of nebulin bound to actin filament in skeletal sarcomere | | Descriptor: | ACTS protein, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Wang, Z, Grange, M, Pospich, S, Wagner, T, Kho, A.L, Gautel, M, Raunser, S. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin.
Science, 375, 2022
|
|
