7X8A
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-11 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7A
 
 | | Cryo-EM structure of SbCas7-11 in complex with crRNA and target RNA | | Descriptor: | RAMP superfamily protein, RNA (33-MER), ZINC ION | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7XC7
 
 | | Cryo-EM structure of a bacterial protein complex | | Descriptor: | CHAT domain protein, RAMP superfamily protein, RNA (33-MER), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7X7R
 
 | | Cryo-EM structure of a bacterial protein | | Descriptor: | RAMP superfamily protein, RNA (36-MER), RNA (5'-R(P*AP*GP*UP*CP*CP*GP*GP*GP*GP*CP*AP*GP*AP*AP*AP*AP*UP*UP*GP*G)-3'), ... | | Authors: | Yu, G, Wang, X, Deng, Z, Zhang, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and function of a bacterial type III-E CRISPR-Cas7-11 complex.
Nat Microbiol, 7, 2022
|
|
7YCX
 
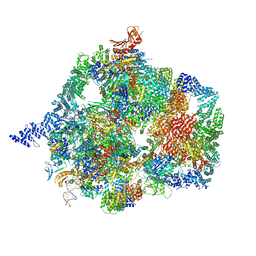 | | The structure of INTAC-PEC complex | | Descriptor: | DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, DNA-directed RNA polymerase II subunit RPB1,DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Zheng, H, Jin, Q, Wang, X, Qi, Y, Liu, W, Ren, Y, Zhao, D, Chen, F.X, Cheng, J, Chen, X, Xu, Y. | | Deposit date: | 2022-07-02 | | Release date: | 2023-03-15 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Structural basis of INTAC-regulated transcription.
Protein Cell, 14, 2023
|
|
7VYV
 
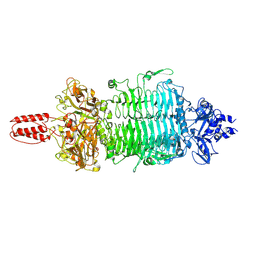 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
7VZ3
 
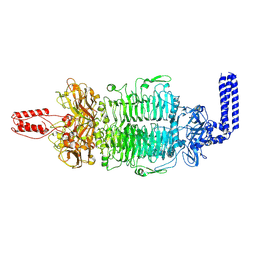 | | Cryo-EM structure of Depo32, a Klebsiella phage depolymerase targets the K2 serotype K. pneumoniae | | Descriptor: | Depolymerase | | Authors: | Cai, R, Ren, Z, Zhao, R, Wang, X, Guo, Z, Du, R, Han, W, Ru, H, Gu, J. | | Deposit date: | 2021-11-15 | | Release date: | 2023-08-30 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural biology and functional features of phage-derived depolymerase Depo32 on Klebsiella pneumoniae with K2 serotype capsular polysaccharides.
Microbiol Spectr, 11, 2023
|
|
7YF3
 
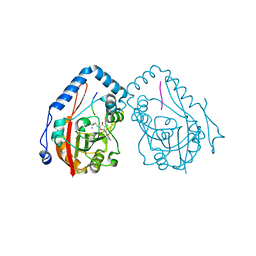 | |
7YF4
 
 | |
7Y9C
 
 | |
7YF2
 
 | |
7CTE
 
 | | Human Origin Recognition Complex, ORC2-5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 2, Origin recognition complex subunit 3, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTF
 
 | | Human origin recognition complex 1-5 State II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTG
 
 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7DHD
 
 | | Vibrio vulnificus Wzb | | Descriptor: | CHLORIDE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7DHF
 
 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | GLYCEROL, PHOSPHATE ION, Protein-tyrosine-phosphatase | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.211 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7DHE
 
 | | Vibrio vulnificus Wzb in complex with benzylphosphonate | | Descriptor: | Protein-tyrosine-phosphatase, benzylphosphonic acid | | Authors: | Ma, Q, Wang, X. | | Deposit date: | 2020-11-14 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Wzb of Vibrio vulnificus represents a new group of low-molecular-weight protein tyrosine phosphatases with a unique insertion in the W-loop.
J.Biol.Chem., 296, 2021
|
|
7D5Z
 
 | | Crystal structure of EBV gH/gL bound with neutralizing antibody 1D8 | | Descriptor: | Envelope glycoprotein H, Envelope glycoprotein L, heavy chain of 1D8, ... | | Authors: | Zhu, Q, Shan, S, Yu, J, Wang, X, Zhang, L, Zeng, M. | | Deposit date: | 2020-09-28 | | Release date: | 2021-10-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | A Neutralizing Antibody Targeting a New Site of Vulnerability on Epstein-Barr Virus gH/gL Protects against Dual-Tropic Infection
To Be Published
|
|
7EB2
 
 | | Cryo-EM structure of human GABA(B) receptor-Gi protein complex | | Descriptor: | (3S)-5,7-ditert-butyl-3-oxidanyl-3-(trifluoromethyl)-1-benzofuran-2-one, Gamma-aminobutyric acid type B receptor subunit 1, Gamma-aminobutyric acid type B receptor subunit 2, ... | | Authors: | Shen, C, Mao, C, Xu, C, Jin, N, Zhang, H, Shen, D, Shen, Q, Wang, X, Hou, T, Rondard, P, Chen, Z, Pin, J, Zhang, Y, Liu, J. | | Deposit date: | 2021-03-08 | | Release date: | 2021-05-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of GABA B receptor-G i protein coupling.
Nature, 594, 2021
|
|
7E3N
 
 | | Crystal structure of Isocitrate dehydrogenase D252N mutant from Trypanosoma brucei in complexed with NADP+, alpha-ketoglutarate, and ca2+ | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP], ... | | Authors: | Arai, N, Shiba, T, Inaoka, D.K, Kita, K, Wang, X, Otani, M, Matsushiro, S, Kojima, C. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Isocitrate dehydrogenase from Trypanosoma brucei.
To Be Published
|
|
7XD9
 
 | | Crystal Structure of Dengue Virus serotype 2 (DENV2) Polymerase Elongation Complex (CTP Form) | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7XD8
 
 | | Crystal Structure of Dengue Virus Serotype 2 (DENV2) Polymerase Elongation Complex (Native Form) | | Descriptor: | GLYCEROL, NS5, RNA (30-mer), ... | | Authors: | Wu, J, Wang, X, Gong, P. | | Deposit date: | 2022-03-26 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of transition from initiation to elongation in de novo viral RNA-dependent RNA polymerases.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5X45
 
 | | Crystal structure of 2A protease from Human rhinovirus C15 | | Descriptor: | ZINC ION, protease 2A | | Authors: | Ling, H, Yang, P, Shaw, N, Sun, Y, Wang, X. | | Deposit date: | 2017-02-10 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | Structural view of the 2A protease from human rhinovirus C15.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
7F7H
 
 | | SARS-CoV-2 S protein RBD in complex with A8-1 Fab | | Descriptor: | Heavy chain of A8-1 Fab, Light chain of A8-1 Fab, Spike glycoprotein S1 | | Authors: | Dou, Y, Wang, X, Wang, K, Liu, P, Lu, B. | | Deposit date: | 2021-06-29 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | High throughput isolation of potent neutralizing antibodies from convalescent COVID-19 patients.
To Be Published
|
|
2K1H
 
 | | Solution NMR structure of SeR13 from Staphylococcus epidermidis. Northeast Structural Genomics Consortium target SeR13 | | Descriptor: | Uncharacterized protein SeR13 | | Authors: | Lee, H, Wylie, G, Bansal, S, Wang, X, Shastry, R, Jiang, M, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-05 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of SeR13.
To be Published
|
|
