1FXL
 
 | |
1G2E
 
 | |
1BML
 
 | |
6KEA
 
 | | crystal structure of MBP-tagged REV7-IpaB complex | | Descriptor: | Maltose-binding periplasmic protein,LINKER,hREV7,LINKER,Invasin IpaB,hREV3 | | Authors: | Wang, X, Pernicone, N, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2019-07-04 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
5IL1
 
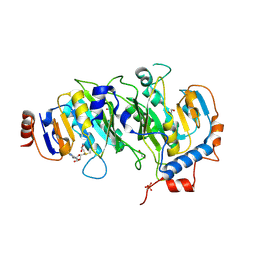 | | Crystal structure of SAM-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL0
 
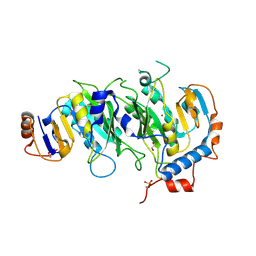 | | Crystal structural of the METTL3-METTL14 complex for N6-adenosine methylation | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, METTL14, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
5IL2
 
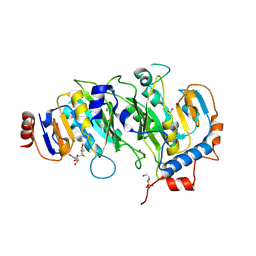 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
7A1V
 
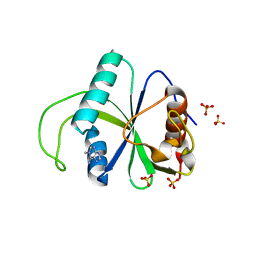 | |
1IH0
 
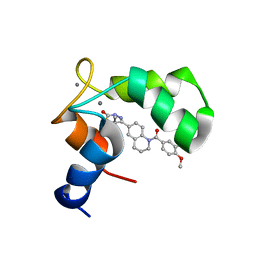 | | Structure of the C-domain of Human Cardiac Troponin C in Complex with Ca2+ Sensitizer EMD 57033 | | Descriptor: | 5-[1-(3,4-DIMETHOXY-BENZOYL)-1,2,3,4-TETRAHYDRO-QUINOLIN-6-YL]-6-METHYL-3,6-DIHYDRO-[1,3,4]THIADIAZIN-2-ONE, CALCIUM ION, TROPONIN C, ... | | Authors: | Wang, X, Li, M.X, Spyracopoulos, L, Beier, N, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the C-domain of human cardiac troponin C in complex with the Ca2+ sensitizing drug EMD 57033.
J.Biol.Chem., 276, 2001
|
|
1JWP
 
 | |
6M3C
 
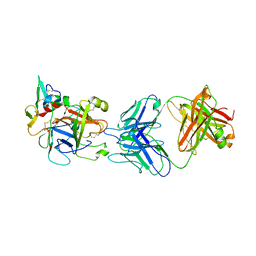 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
4QPG
 
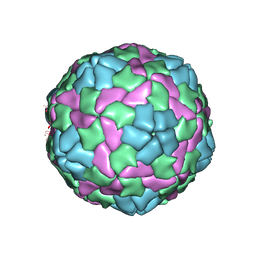 | | Crystal structure of empty hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP0, Capsid protein VP1, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
4QPI
 
 | | Crystal structure of hepatitis A virus | | Descriptor: | CHLORIDE ION, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Wang, X, Ren, J, Gao, Q, Hu, Z, Sun, Y, Li, X, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2014-06-23 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Hepatitis A virus and the origins of picornaviruses.
Nature, 517, 2015
|
|
1C4P
 
 | | BETA DOMAIN OF STREPTOKINASE | | Descriptor: | PROTEIN (STREPTOKINASE) | | Authors: | Wang, X, Zhang, X.C. | | Deposit date: | 1999-09-15 | | Release date: | 1999-10-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of streptokinase beta-domain.
FEBS Lett., 459, 1999
|
|
1NYM
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (CXB) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NYY
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1NY0
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (NBF) | | Descriptor: | Beta-lactamase TEM, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1JVJ
 
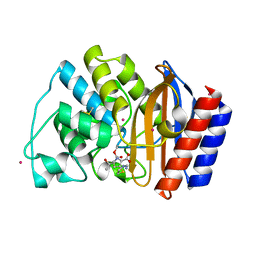 | | CRYSTAL STRUCTURE OF N132A MUTANT OF TEM-1 BETA-LACTAMASE IN COMPLEX WITH A N-FORMIMIDOYL-THIENAMYCINE | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, BETA-LACTAMASE TEM, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2001-08-30 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Noncovalent interaction energies in covalent complexes: TEM-1 beta-lactamase and beta-lactams.
Proteins, 47, 2002
|
|
1JWZ
 
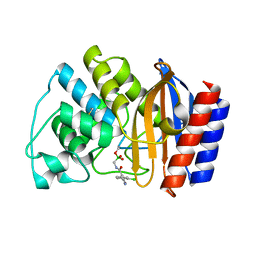 | |
1JWV
 
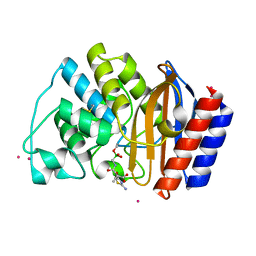 | |
1LI0
 
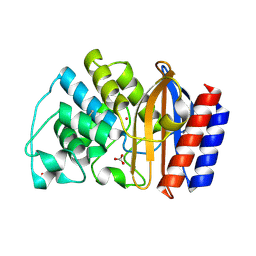 | | Crystal structure of TEM-32 beta-Lactamase at 1.6 Angstrom | | Descriptor: | BICARBONATE ION, Class A beta-Lactamase- TEM-32, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
1IB2
 
 | |
1LHY
 
 | | Crystal structure of TEM-30 beta-Lactamase at 2.0 Angstrom | | Descriptor: | Class A beta-Lactamase- TEM 30, PHOSPHATE ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
1NXY
 
 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (SM2) | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-(3-CARBOXYPHENYL)METHYLBORONIC ACID, Beta-lactamase TEM, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-11 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition and Resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1LI9
 
 | | Crystal structure of TEM-34 beta-Lactamase at 1.5 Angstrom | | Descriptor: | Class A beta-Lactamase- TEM-34, PHOSPHATE ION, POTASSIUM ION | | Authors: | Wang, X, Minasov, G, Shoichet, B.K. | | Deposit date: | 2002-04-17 | | Release date: | 2002-09-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The structural bases of antibiotic resistance in the clinically derived mutant beta-lactamases TEM-30, TEM-32, and TEM-34.
J.Biol.Chem., 277, 2002
|
|
