1R0M
 
 | | Structure of Deinococcus radiodurans N-acylamino acid racemase at 1.3 : insights into a flexible binding pocket and evolution of enzymatic activity | | Descriptor: | N-acylamino acid racemase | | Authors: | Wang, W.-C, Chiu, W.-C, Hsu, S.-K, Wu, C.-L, Chen, C.-Y, Liu, J.-S, Hsu, W.-H. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis for catalytic racemization and substrate specificity of an N-acylamino acid racemase homologue from Deinococcus radiodurans
J.Mol.Biol., 342, 2004
|
|
5X0I
 
 | |
7RAX
 
 | |
8DVR
 
 | | Cryo-EM structure of RIG-I bound to the end of p3SLR30 (+AMPPNP) | | Descriptor: | Antiviral innate immune response receptor RIG-I, GUANOSINE-5'-TRIPHOSPHATE, ZINC ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVS
 
 | | Cryo-EM structure of RIG-I bound to the end of OHSLR30 (+ATP) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Antiviral innate immune response receptor RIG-I, MAGNESIUM ION, ... | | Authors: | Wang, W, Pyle, A.M. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
8DVU
 
 | |
8XZG
 
 | | Cryo-EM structure of the [Pyr1]-apelin-13-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Apelin-13, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZF
 
 | | Cryo-EM structure of the WN561-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZJ
 
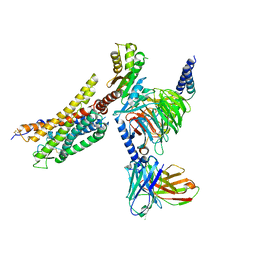 | | Cryo-EM structure of the WN353-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZI
 
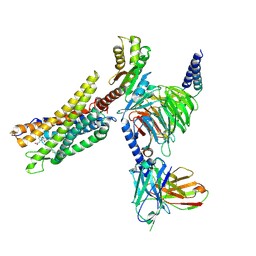 | | Cryo-EM structure of the CMF-019-bound human APLNR-Gi complex | | Descriptor: | (3~{S})-5-methyl-3-[[1-pentan-3-yl-2-(thiophen-2-ylmethyl)benzimidazol-5-yl]carbonylamino]hexanoic acid, Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
8XZH
 
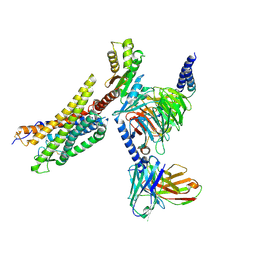 | | Cryo-EM structure of the MM07-bound human APLNR-Gi complex | | Descriptor: | Apelin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, W, Ji, S, Zhang, Y. | | Deposit date: | 2024-01-21 | | Release date: | 2024-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure-based design of non-hypertrophic apelin receptor modulator.
Cell, 187, 2024
|
|
4Y05
 
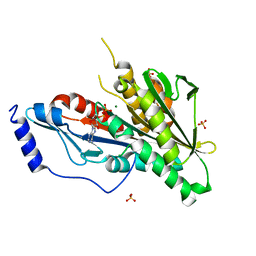 | | KIF2C short Loop2 construct | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2C, MAGNESIUM ION, ... | | Authors: | Wang, W, Knossow, M, Gigant, B. | | Deposit date: | 2015-02-05 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | New Insights into the Coupling between Microtubule Depolymerization and ATP Hydrolysis by Kinesin-13 Protein Kif2C.
J.Biol.Chem., 290, 2015
|
|
5WVM
 
 | | Crystal structure of baeS cocrystallized with 2 mM indole | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Rang, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
5WVN
 
 | | Crystal structure of MBS-BaeS fusion protein | | Descriptor: | Maltose-binding periplasmic protein,Two-component system sensor kinase, SULFATE ION | | Authors: | Wang, W, Zhang, Y, Ran, T, Xu, D. | | Deposit date: | 2016-12-26 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the sensor domain of BaeS from Serratia marcescens FS14
Proteins, 85, 2017
|
|
8YSV
 
 | | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, 6-phospho-beta-glucosidase, ETHANOL, ... | | Authors: | Wang, W.Y, Li, Y.J, Liu, Z.Y, Han, X.D. | | Deposit date: | 2024-03-23 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal structure of beta - glucosidase 6PG from Enterococcus faecalis
To Be Published
|
|
9IZN
 
 | | Crystal structure of HKU1A RBD bound to TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, Transmembrane protease serine 2 | | Authors: | Wang, W, Xu, Y, Zhang, S. | | Deposit date: | 2024-08-01 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure of human coronavirus HKU1 receptor binding domain bound to TMPRSS2 receptor
Hlife, 2024
|
|
5MIO
 
 | | KIF2C-DARPIN FUSION PROTEIN BOUND TO TUBULIN | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF2C,KIF2C FUSED TO A DARPIN,KIF2C FUSED TO A DARPIN, ... | | Authors: | Wang, W, Gigant, B. | | Deposit date: | 2016-11-28 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule disassembly by kinesin-13s from the structure of Kif2C bound to tubulin.
Nat Commun, 8, 2017
|
|
2DYV
 
 | |
2DYU
 
 | |
2E2K
 
 | |
2E2L
 
 | |
3CLH
 
 | | Crystal structure of 3-dehydroquinate synthase (DHQS)from Helicobacter pylori | | Descriptor: | 3-dehydroquinate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Wang, W.C, Liu, J.S, Cheng, W.C, Wang, H.J, Chen, Y.C. | | Deposit date: | 2008-03-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based inhibitor discovery of Helicobacter pylori dehydroquinate synthase.
Biochem.Biophys.Res.Commun., 373, 2008
|
|
3DBQ
 
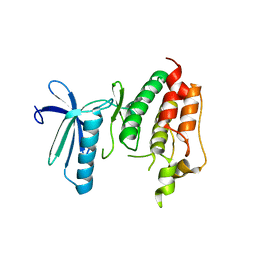 | | Crystal structure of TTK kinase domain | | Descriptor: | Dual specificity protein kinase TTK | | Authors: | Wang, W, Yang, Y.T, Gao, Y.F, Zhu, S.C, Wang, F, Old, W, Xu, Q.B, Resing, K, Ahn, N, Lei, M, Liu, X.D. | | Deposit date: | 2008-06-02 | | Release date: | 2009-02-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Mechanistic Insights into Mps1 Kinase Activation
J.CELL.MOL.MED., 13, 2008
|
|
7XZ4
 
 | |
4DQQ
 
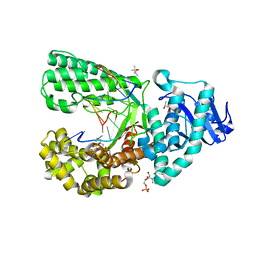 | | Ternary complex of Bacillus DNA Polymerase I Large Fragment E658A, DNA duplex, and rCTP (paired with dG of template) in presence of Mg2+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Wang, W, Beese, L.S. | | Deposit date: | 2012-02-16 | | Release date: | 2012-06-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.595 Å) | | Cite: | Structural factors that determine selectivity of a high fidelity DNA polymerase for deoxy-, dideoxy-, and ribonucleotides.
J.Biol.Chem., 287, 2012
|
|
