4QG9
 
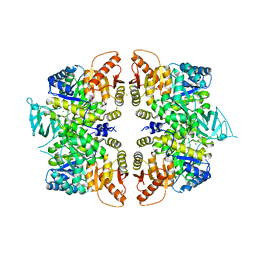 | | crystal structure of PKM2-R399E mutant | | 分子名称: | ACETATE ION, MAGNESIUM ION, Pyruvate kinase PKM | | 著者 | Wang, P, Sun, C, Zhu, T, Xu, Y. | | 登録日 | 2014-05-22 | | 公開日 | 2015-02-25 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.381 Å) | | 主引用文献 | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
4K2X
 
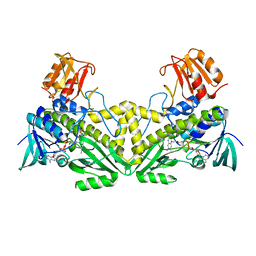 | |
5YUF
 
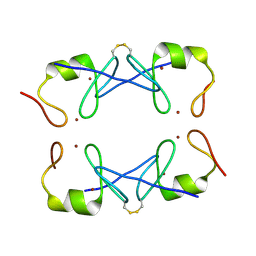 | | Crystal Structure of PML RING tetramer | | 分子名称: | Protein PML, ZINC ION | | 著者 | Wang, P, Benhend, S, Wu, H, Breitenbach, V, Zhen, T, Jollivet, F, Peres, L, Li, Y, Chen, S, Chen, Z, de THE, H, Meng, G. | | 登録日 | 2017-11-22 | | 公開日 | 2018-04-11 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | RING tetramerization is required for nuclear body biogenesis and PML sumoylation.
Nat Commun, 9, 2018
|
|
5U2U
 
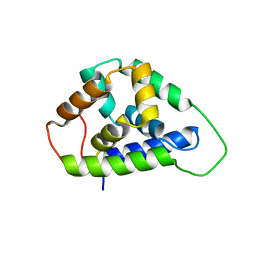 | |
5SV7
 
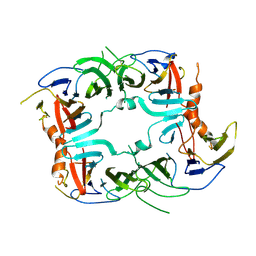 | | The Crystal structure of a chaperone | | 分子名称: | Eukaryotic translation initiation factor 2-alpha kinase 3 | | 著者 | Wang, P, Li, J, Sha, B. | | 登録日 | 2016-08-04 | | 公開日 | 2017-03-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.209 Å) | | 主引用文献 | The ER stress sensor PERK luminal domain functions as a molecular chaperone to interact with misfolded proteins.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
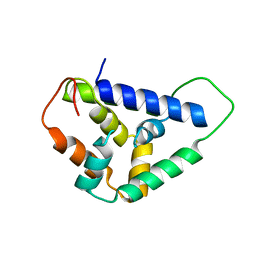
5U2L
 
 | |
5V1D
 
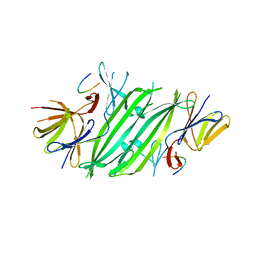 | |
5HOT
 
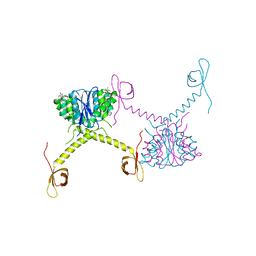 | | Structural Basis for Inhibitor-Induced Aggregation of HIV-1 Integrase | | 分子名称: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, Integrase | | 著者 | Gupta, K, Turkki, V, Sherrill-Mix, S, Hwang, Y, Eilers, G, Taylor, L, McDanal, C, Wang, P, Temelkoff, D, Nolte, R, Velthuisen, E, Jeffrey, J, Van Duyne, G.D, Bushman, F.D. | | 登録日 | 2016-01-19 | | 公開日 | 2016-12-14 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (4.4 Å) | | 主引用文献 | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
1IU0
 
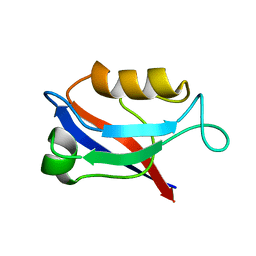 | | The first PDZ domain of PSD-95 | | 分子名称: | PSD-95 | | 著者 | Long, J.-F, Tochio, H, Wang, P, Sala, C, Niethammer, M, Sheng, M, Zhang, M. | | 登録日 | 2002-02-18 | | 公開日 | 2003-03-11 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Supramodular structure and synergistic target binding of the N-terminal tandem PDZ domains of PSD-95
J.MOL.BIOL., 327, 2003
|
|
8HIC
 
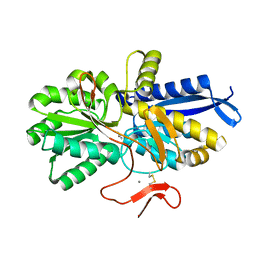 | | Crystal structure of UrtA from Prochlorococcus marinus str. MIT 9313 in complex with urea and calcium | | 分子名称: | CALCIUM ION, Putative urea ABC transporter, substrate binding protein, ... | | 著者 | Zhang, Y.Z, Wang, P, Wang, C. | | 登録日 | 2022-11-19 | | 公開日 | 2023-11-22 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structural and molecular basis for urea recognition by Prochlorococcus.
J.Biol.Chem., 299, 2023
|
|
8H0Z
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-122 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
6AK1
 
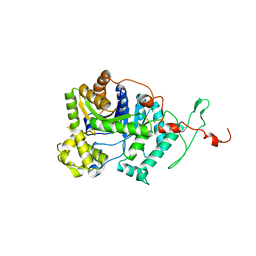 | | Crystal structure of DmoA from Hyphomicrobium sulfonivorans | | 分子名称: | Dimethyl-sulfide monooxygenase | | 著者 | Cao, H.Y, Wang, P, Peng, M, Li, C.Y. | | 登録日 | 2018-08-28 | | 公開日 | 2018-12-12 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.284 Å) | | 主引用文献 | Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | 分子名称: | E3 ubiquitin-protein ligase UHRF1, Spacer | | 著者 | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | 登録日 | 2016-02-22 | | 公開日 | 2016-04-20 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
3NK6
 
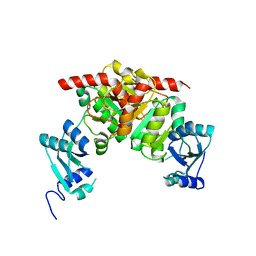 | | Structure of the Nosiheptide-resistance methyltransferase | | 分子名称: | 23S rRNA methyltransferase | | 著者 | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | 登録日 | 2010-06-18 | | 公開日 | 2010-07-21 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
4WQN
 
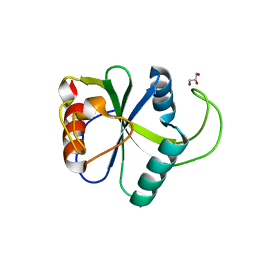 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | 分子名称: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | 著者 | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | 登録日 | 2014-10-22 | | 公開日 | 2014-11-19 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.121 Å) | | 主引用文献 | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
3NK7
 
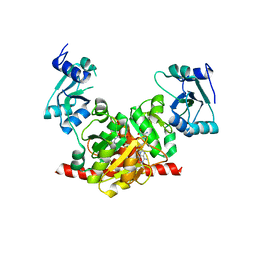 | | Structure of the Nosiheptide-resistance methyltransferase S-adenosyl-L-methionine Complex | | 分子名称: | 23S rRNA methyltransferase, S-ADENOSYLMETHIONINE | | 著者 | Yang, H, Wang, Z, Shen, Y, Wang, P, Murchie, A, Xu, Y. | | 登録日 | 2010-06-18 | | 公開日 | 2010-07-21 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal Structure of the Nosiheptide-Resistance Methyltransferase of Streptomyces actuosus
Biochemistry, 49, 2010
|
|
6IZT
 
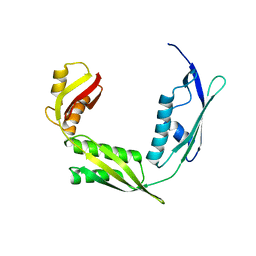 | | Crystal structure of Haemophilus Influenzae BamA POTRA3-5 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6IZS
 
 | | Crystal structure of Haemophilus influenzae BamA POTRA4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-20 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
6J09
 
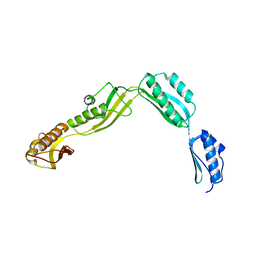 | | Crystal structure of Haemophilus Influenzae BamA POTRA1-4 | | 分子名称: | Outer membrane protein assembly factor BamA | | 著者 | Ma, X, Wang, Q, Li, Y, Tan, P, Wu, H, Wang, P, Dong, X, Hong, L, Meng, G. | | 登録日 | 2018-12-21 | | 公開日 | 2019-10-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | How BamA recruits OMP substratesviapoly-POTRAs domain.
Faseb J., 33, 2019
|
|
3PUR
 
 | | CEKDM7A from C.Elegans, complex with D-2-HG | | 分子名称: | (2R)-2-hydroxypentanedioic acid, FE (II) ION, Lysine-specific demethylase 7 homolog, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
3PUQ
 
 | | CEKDM7A from C.Elegans, complex with alpha-KG | | 分子名称: | 2-OXOGLUTARIC ACID, FE (II) ION, GLYCEROL, ... | | 著者 | Yang, Y, Wang, P, Xu, W, Xu, Y. | | 登録日 | 2010-12-06 | | 公開日 | 2011-01-26 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases
Cancer Cell, 19, 2011
|
|
8H13
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x2 Disulfide (G400C and V969C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (4.05 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H10
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Locked-2 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.99 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H14
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x3 Disulfide (D414C and V969C), Locked-1 Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-10-19 | | 最終更新日 | 2024-10-09 | | 実験手法 | ELECTRON MICROSCOPY (3.39 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
8H11
 
 | | Structure of SARS-CoV-1 Spike Protein with Engineered x1 Disulfide (S370C and D967C), Closed Conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | 著者 | Zhang, X, Li, Z, Liu, Y, Wang, J, Fu, L, Wang, P, He, J, Xiong, X. | | 登録日 | 2022-09-30 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-07-19 | | 実験手法 | ELECTRON MICROSCOPY (2.72 Å) | | 主引用文献 | Disulfide stabilization reveals conserved dynamic features between SARS-CoV-1 and SARS-CoV-2 spikes.
Life Sci Alliance, 6, 2023
|
|
