5GXF
 
 | |
5GXE
 
 | |
4BFC
 
 | | Crystal structure of the C-terminal CMP-Kdo binding domain of WaaA from Acinetobacter baumannii | | Descriptor: | 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE, BETA-MERCAPTOETHANOL, SULFATE ION | | Authors: | Kimbung, Y.R, Hakansson, M, Logan, D, Wang, P.F, Schulz, M, Mamat, U, Woodard, R.W. | | Deposit date: | 2013-03-18 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the C-Terminal Cmp-Kdo Binding Domain of Waaa from Acinetobacter Baumannii
To be Published
|
|
4NM6
 
 | | Crystal structure of TET2-DNA complex | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*T)-3', FE (II) ION, Methylcytosine dioxygenase TET2, ... | | Authors: | Hu, L, Li, Z, Cheng, J, Rao, Q, Gong, W, Liu, M, Wang, P, Xu, Y. | | Deposit date: | 2013-11-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.026 Å) | | Cite: | Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation.
Cell(Cambridge,Mass.), 155, 2013
|
|
2JYT
 
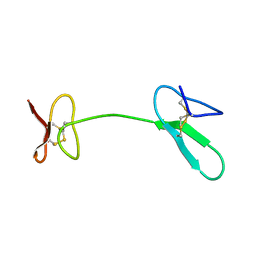 | | Human Granulin C, isomer 1 | | Descriptor: | Granulin-5 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
2JYU
 
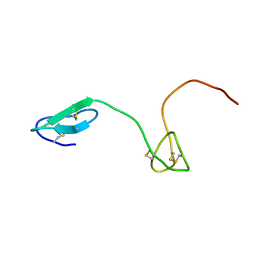 | | Human Granulin C, isomer 2 | | Descriptor: | Granulin-5 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
2JYV
 
 | | Human Granulin F | | Descriptor: | Granulin-2 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
2JYE
 
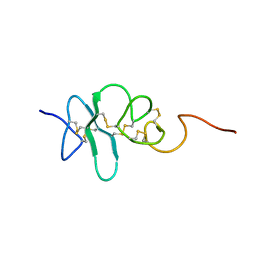 | | Human Granulin A | | Descriptor: | Granulin A | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-13 | | Release date: | 2008-04-22 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
7KPL
 
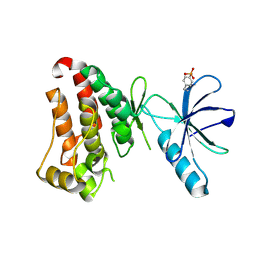 | | Crystal structure of hEphB1 in apo form | | Descriptor: | Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KPM
 
 | | Crystal structure of hEphB1 bound with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ephrin type-B receptor 1 | | Authors: | Ahmed, M, Wang, P, Sadek, H. | | Deposit date: | 2020-11-11 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.608 Å) | | Cite: | Identification of tetracycline combinations as EphB1 tyrosine kinase inhibitors for treatment of neuropathic pain.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6CSD
 
 | | V308E mutant of cytochrome P450 2D6 complexed with prinomastat | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Yang, Y.T, Fujita, K, Wang, P.F, Im, S.C, Pearl, N.M, Meagher, J, Stuckey, J, Waskell, L. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Characteristic conformational changes on the distal and proximal surfaces of cytochrome P450 2D6 in response to substrate binding
To Be Published
|
|
6CSB
 
 | | V308E mutant of cytochrome P450 2D6 complexed with thioridazine | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, ACETATE ION, Cytochrome P450 2D6, ... | | Authors: | Yang, Y.T, Fujita, K, Wang, P.F, Im, S.C, Pearl, N.M, Meagher, J, Stuckey, J, Waskell, L. | | Deposit date: | 2018-03-20 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Characteristic conformational changes on the distal and proximal surfaces of cytochrome P450 2D6 in response to substrate binding
To Be Published
|
|
6IMQ
 
 | | Crystal structure of PML B1-box multimers | | Descriptor: | CHLORIDE ION, Protein PML, ZINC ION | | Authors: | Li, Y, Ma, X, Chen, Z, Wu, H, Wang, P, Wu, W, Cheng, N, Zeng, L, Zhang, H, Cai, X, Chen, S.J, Chen, Z, Meng, G. | | Deposit date: | 2018-10-23 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | B1 oligomerization regulates PML nuclear body biogenesis and leukemogenesis.
Nat Commun, 10, 2019
|
|
3RUC
 
 | |
3RUE
 
 | |
3RUA
 
 | |
3RUF
 
 | |
3RU9
 
 | |
3RUH
 
 | |
3RUD
 
 | |
7F8T
 
 | | Re-refinement of the 2XRY X-ray structure of archaeal class II CPD photolyase from Methanosarcina mazei | | Descriptor: | Deoxyribodipyrimidine photolyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-07-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
5Z16
 
 | |
3PEL
 
 | | Structure of Greyhound Hemoglobin: Origin of High Oxygen Affinity | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bhatt, V.S, Zaldivar-Lopez, S, Harris, D.R, Couto, C.G, Wang, P.G, Palmer, A.F. | | Deposit date: | 2010-10-26 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Greyhound hemoglobin: origin of high oxygen affinity.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
6IJB
 
 | | Structure of 3-methylmercaptopropionate CoA ligase mutant K523A in complex with AMP and MMPA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(methylsulfanyl)propanoic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
