8GQU
 
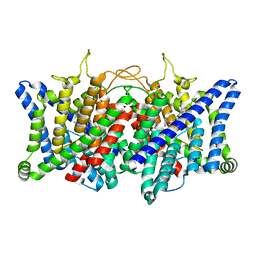 | | AK-42 inhibitor binding human ClC-2 TMD | | 分子名称: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | 著者 | Wang, L. | | 登録日 | 2022-08-30 | | 公開日 | 2023-07-05 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
7TAI
 
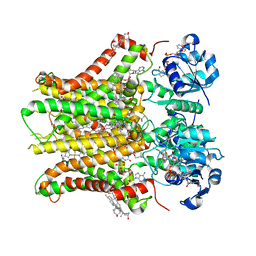 | | Structure of STEAP2 in complex with ligands | | 分子名称: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Wang, L, Chen, K.H, Zhou, M. | | 登録日 | 2021-12-20 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-02-07 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Mechanism of stepwise electron transfer in six-transmembrane epithelial antigen of the prostate (STEAP) 1 and 2.
Elife, 12, 2023
|
|
1BLR
 
 | | NMR SOLUTION STRUCTURE OF HUMAN CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II, 22 STRUCTURES | | 分子名称: | CELLULAR RETINOIC ACID BINDING PROTEIN-TYPE II | | 著者 | Wang, L, Li, Y, Abilddard, F, Yan, H, Markely, J. | | 登録日 | 1998-07-20 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR solution structure of type II human cellular retinoic acid binding protein: implications for ligand binding.
Biochemistry, 37, 1998
|
|
6JFI
 
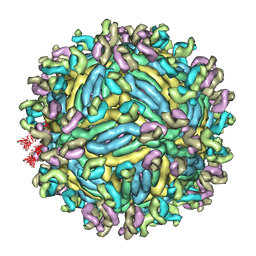 | | The symmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | 分子名称: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural protein E, ... | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (11 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JFH
 
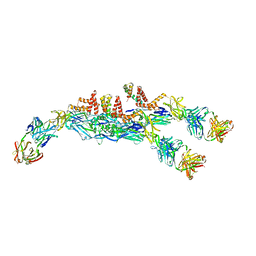 | | The asymmetric-reconstructed cryo-EM structure of Zika virus-FabZK2B10 complex | | 分子名称: | FabZK2B10 heavy chain, FabZK2B10 light chain, ZIKV structural E protein, ... | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-08 | | 公開日 | 2019-04-10 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (20 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
6JEP
 
 | | Structure of a neutralizing antibody bound to the Zika envelope protein domain III | | 分子名称: | Genome polyprotein, heavy chain of Fab ZK2B10, light chain of Fab ZK2B10 | | 著者 | Wang, L, Wang, R.K, Wang, L, Ben, H.J, Yu, L, Gao, F, Shi, X.L, Yin, C.B, Zhang, F.C, Xiang, Y, Zhang, L.Q. | | 登録日 | 2019-02-07 | | 公開日 | 2019-05-15 | | 最終更新日 | 2019-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.316 Å) | | 主引用文献 | Structural Basis for Neutralization and Protection by a Zika Virus-Specific Human Antibody.
Cell Rep, 26, 2019
|
|
3Q4A
 
 | |
3MCE
 
 | | Crystal structure of the NAC domain of alpha subunit of nascent polypeptide-associated complex(NAC) | | 分子名称: | IODIDE ION, Nascent polypeptide-associated complex subunit alpha | | 著者 | Wang, L.F, Zhang, W.C, Wang, L, Zhang, X.J.C, Li, X.M, Rao, Z. | | 登録日 | 2010-03-29 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.396 Å) | | 主引用文献 | Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit
Protein Cell, 1, 2010
|
|
7U4T
 
 | | Human V-ATPase in state 2 with SidK and mEAK-7 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MTOR-associated protein MEAK7, ... | | 著者 | Wang, L, Fu, T.M. | | 登録日 | 2022-03-01 | | 公開日 | 2022-08-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Identification of mEAK-7 as a human V-ATPase regulator via cryo-EM data mining.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
4YPR
 
 | | Crystal Structure of D144N MutY bound to its anti-substrate | | 分子名称: | A/G-specific adenine glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), DNA (5'-D(*T*GP*TP*CP*CP*AP*CP*GP*TP*CP*T)-3'), ... | | 著者 | Wang, L, Lee, S, Verdine, G.L. | | 登録日 | 2015-03-13 | | 公開日 | 2015-05-27 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structural Basis for Avoidance of Promutagenic DNA Repair by MutY Adenine DNA Glycosylase.
J.Biol.Chem., 290, 2015
|
|
7YR4
 
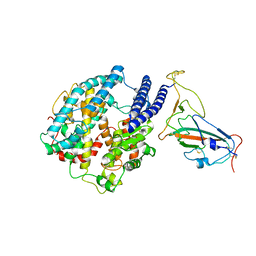 | |
7RR3
 
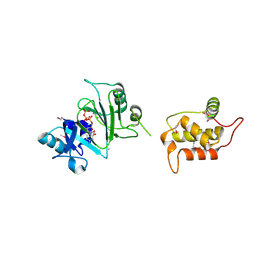 | | Structure of Deep-Sea Phage NrS-1 Primase-Polymerase N300 in complex with calcium and ddCTP | | 分子名称: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, CALCIUM ION, Primase | | 著者 | Wang, L, Yu, C, Sliz, P. | | 登録日 | 2021-08-09 | | 公開日 | 2022-01-12 | | 最終更新日 | 2022-01-19 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Molecular Dissection of the Primase and Polymerase Activities of Deep-Sea Phage NrS-1 Primase-Polymerase.
Front Microbiol, 12, 2021
|
|
7RR4
 
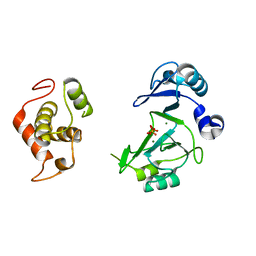 | |
7UPT
 
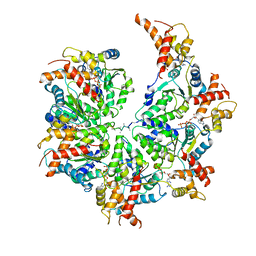 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (open conformation) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | 著者 | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | 登録日 | 2022-04-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
7UPR
 
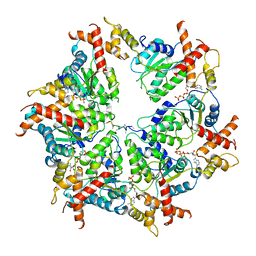 | | Human mitochondrial AAA protein ATAD1 (with a catalytic dead mutation) in complex with a peptide substrate (closed conformation) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Outer mitochondrial transmembrane helix translocase, ... | | 著者 | Wang, L, Toutkoushian, H, Belyy, V, Kokontis, C, Walter, P. | | 登録日 | 2022-04-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | Conserved structural elements specialize ATAD1 as a membrane protein extraction machine.
Elife, 11, 2022
|
|
3MCB
 
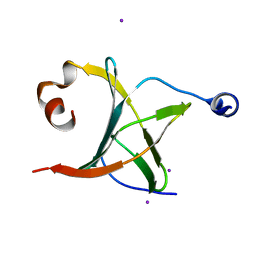 | | Crystal structure of NAC domains of human nascent polypeptide-associated complex (NAC) | | 分子名称: | IODIDE ION, Nascent polypeptide-associated complex subunit alpha, Transcription factor BTF3 | | 著者 | Wang, L.F, Zhang, W.C, Wang, L, Zhang, X.J.C, Li, X.M, Rao, Z. | | 登録日 | 2010-03-29 | | 公開日 | 2010-07-14 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit
Protein Cell, 1, 2010
|
|
5KN9
 
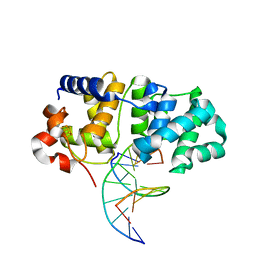 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | 分子名称: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | 著者 | Wang, L, Chakravarthy, S, Verdine, G.L. | | 登録日 | 2016-06-27 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5KN8
 
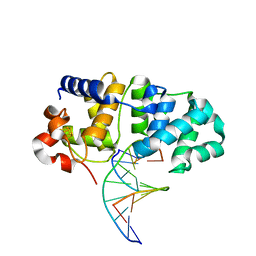 | | MutY N-terminal domain in complex with undamaged DNA | | 分子名称: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | 著者 | Wang, L, Chakravarthy, S, Verdine, G.L. | | 登録日 | 2016-06-27 | | 公開日 | 2017-02-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
7TYJ
 
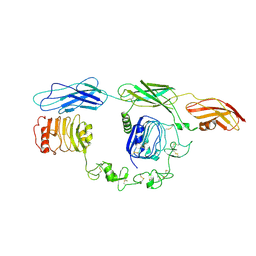 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was focused on one of two halves with C1 symmetry applied | | 分子名称: | Insulin receptor-related protein | | 著者 | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYK
 
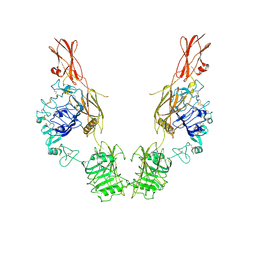 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in apo-state captured at pH 7. The 3D refinement was applied with C2 symmetry | | 分子名称: | Insulin receptor-related protein | | 著者 | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7TYM
 
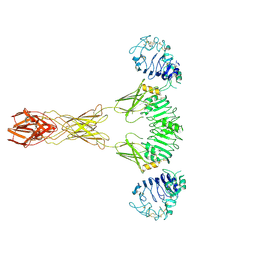 | | Cryo-EM Structure of insulin receptor-related receptor (IRR) in active-state captured at pH 9. The 3D refinement was applied with C2 symmetry | | 分子名称: | Insulin receptor-related protein | | 著者 | Wang, L.W, Hall, C, Li, J, Choi, E, Bai, X.C. | | 登録日 | 2022-02-13 | | 公開日 | 2023-02-15 | | 最終更新日 | 2023-05-31 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Structural basis of the alkaline pH-dependent activation of insulin receptor-related receptor.
Nat.Struct.Mol.Biol., 30, 2023
|
|
6FXD
 
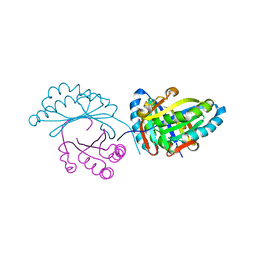 | | Crystal structure of MupZ from Pseudomonas fluorescens | | 分子名称: | MupZ | | 著者 | Wang, L, Parnell, A, Williams, C, Bakar, N.A, van der Kamp, M.W, Simpson, T.J, Race, P.R, Crump, M.P, Willis, C.L. | | 登録日 | 2018-03-08 | | 公開日 | 2019-03-06 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | A Rieske oxygenase/epoxide hydrolase-catalysed reaction cascade creates oxygen heterocycles in mupirocin biosynthesis
Nat Catal, 2018
|
|
3Q47
 
 | |
3Q49
 
 | |
6PE0
 
 | | Msp1 (E214Q)-substrate complex | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Membrane-spanning ATPase-like protein, ... | | 著者 | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | 登録日 | 2019-06-19 | | 公開日 | 2020-02-12 | | 最終更新日 | 2024-03-20 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
