5V2C
 
 | | RE-REFINEMENT OF CRYSTAL STRUCTURE OF PHOTOSYSTEM II COMPLEX | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Wang, J, Wiwczar, J.M, Brudvig, G.W. | | Deposit date: | 2017-03-03 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chlorophyll a with a farnesyl tail in thermophilic cyanobacteria.
Photosyn. Res., 134, 2017
|
|
5VCJ
 
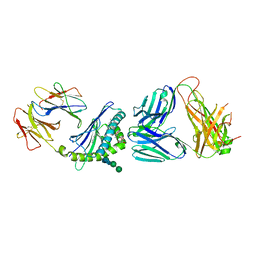 | | Structure of alpha-galactosylphytosphingosine bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (2S,3S,4R)-2-amino-3,4-dihydroxyoctadecyl alpha-D-galactopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, Antigen-presenting glycoprotein CD1d1, ... | | Authors: | Wang, J, Zajonc, D.M. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Enhancing T cell responses and tumour immunity by vaccination with peptides conjugated to a weak NKT cell agonist.
Org. Biomol. Chem., 17, 2019
|
|
3ONG
 
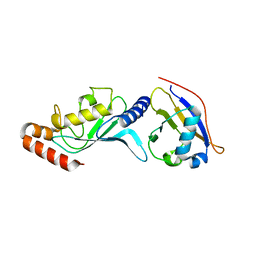 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | Descriptor: | SUMO-conjugating enzyme UBC9, Ubiquitin-activating enzyme E1-like | | Authors: | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | Deposit date: | 2010-08-28 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
3ONH
 
 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | Descriptor: | Ubiquitin-activating enzyme E1-like | | Authors: | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | Deposit date: | 2010-08-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
3PLS
 
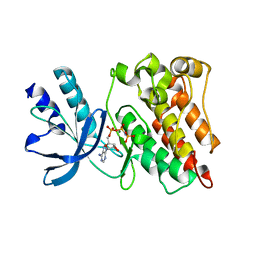 | | RON in complex with ligand AMP-PNP | | Descriptor: | MAGNESIUM ION, Macrophage-stimulating protein receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Wang, J, Steinbacher, S, Augustin, M, Schreiner, P, Epstein, D, Mulvihill, M.J, Crew, A.P. | | Deposit date: | 2010-11-15 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Crystal Structure of a Constitutively Active Mutant RON Kinase Suggests an Intramolecular Autophosphorylation Hypothesis
Biochemistry, 49, 2010
|
|
7CPX
 
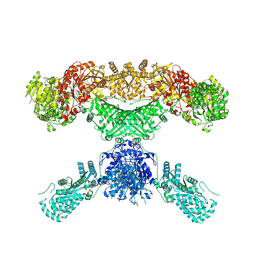 | | Lovastatin nonaketide synthase | | Descriptor: | Lovastatin nonaketide synthase, polyketide synthase component, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
7CPY
 
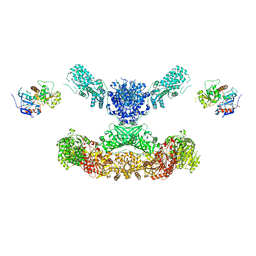 | | Lovastatin nonaketide synthase with LovC | | Descriptor: | Lovastatin nonaketide synthase, enoyl reductase component lovC, polyketide synthase component, ... | | Authors: | Wang, J, Wang, Z. | | Deposit date: | 2020-08-08 | | Release date: | 2021-01-13 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for the biosynthesis of lovastatin.
Nat Commun, 12, 2021
|
|
4EQG
 
 | |
6J5S
 
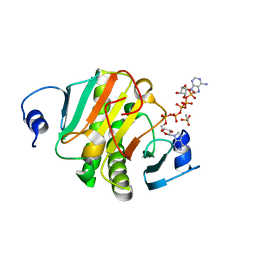 | | Crystal structure of human HINT1 mutant complexing with AP5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J53
 
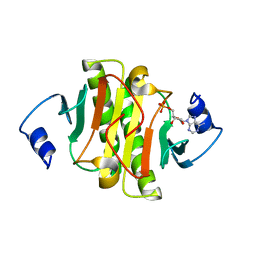 | | Crystal structure of human HINT1 complexing with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J58
 
 | | Crystal structure of human HINT1 complexing with AP4A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J64
 
 | | Crystal structure of human HINT1 mutant complexing with AP4A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J5Z
 
 | | Crystal structure of human HINT1 mutant complexing with AP3A | | Descriptor: | ADENOSINE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-12 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
8HQG
 
 | |
8HXS
 
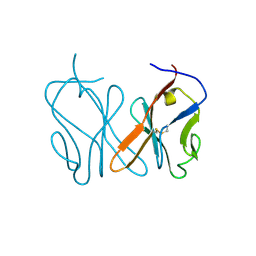 | | Small_spotted catshark CD8alpha | | Descriptor: | T-cell surface glycoprotein CD8 alpha chain | | Authors: | Wang, J, Zou, J. | | Deposit date: | 2023-01-05 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The first crystal structure of CD8 alpha alpha from a cartilaginous fish.
Front Immunol, 14, 2023
|
|
4EQH
 
 | |
8HVM
 
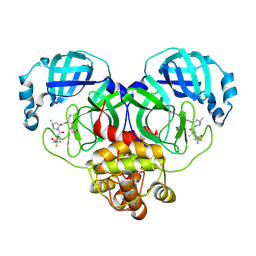 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07321332 | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-27 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of SARS-Cov-2 main protease
K90R mutant in complex with PF07321332
To Be Published
|
|
8HVY
 
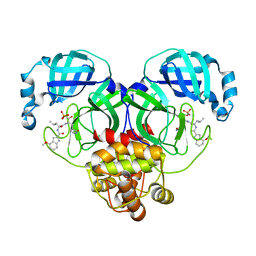 | | Crystal structure of SARS-Cov-2 main protease K90R mutant in complex with PF07304814 | | Descriptor: | 3C-like proteinase nsp5, [(3~{S})-3-[[(2~{S})-2-[(4-methoxy-1~{H}-indol-2-yl)carbonylamino]-4-methyl-pentanoyl]amino]-2-oxidanylidene-4-[(3~{R})-2-oxidanylidene-3,4-dihydropyrrol-3-yl]butyl] dihydrogen phosphate | | Authors: | Wang, J, Zhang, J, Li, J. | | Deposit date: | 2022-12-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structures of main protease (M pro ) mutants of SARS-CoV-2 variants bound to PF-07304814.
Mol Biomed, 4, 2023
|
|
6J65
 
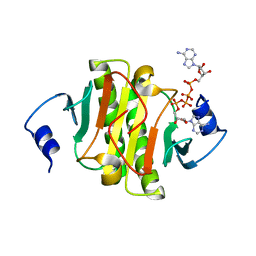 | | Crystal structure of human HINT1 mutant complexing with AP4A II | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
4EQE
 
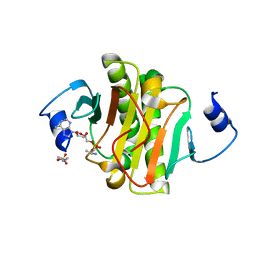 | |
1WAF
 
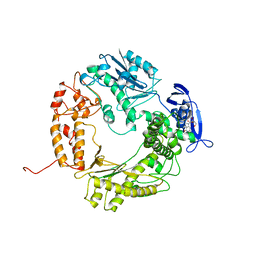 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
1WAJ
 
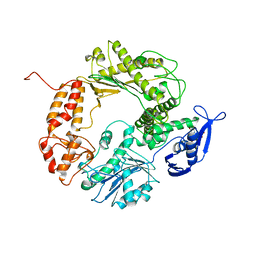 | | DNA POLYMERASE FROM BACTERIOPHAGE RB69 | | Descriptor: | DNA POLYMERASE, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Wang, J, Satter, A.K.M.A, Wang, C.C, Karam, J.D, Konigsberg, W.H, Steitz, T.A. | | Deposit date: | 1997-04-13 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69.
Cell(Cambridge,Mass.), 89, 1997
|
|
3V69
 
 | | Filia-N crystal structure | | Descriptor: | Protein Filia | | Authors: | Wang, J, Xu, M, Zhu, K, Li, L, Liu, X. | | Deposit date: | 2011-12-19 | | Release date: | 2012-02-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The N-terminus of FILIA Forms an Atypical KH Domain with a Unique Extension Involved in Interaction with RNA.
Plos One, 7, 2012
|
|
6K3E
 
 | | LSD1/Co-Rest structure with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PCPA derivative, ... | | Authors: | Wang, J. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | LSD1/Co-Rest structure with an inhibitor
To Be Published
|
|
7LHL
 
 | |
