8XOU
 
 | | Prohead portal vertex of bacteriophage lambda | | Descriptor: | Major capsid protein, Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.58 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XQB
 
 | | Mature virion portal vertex of bacteriophage lambda | | Descriptor: | Capsid decoration protein, Head completion protein, Head-tail connector protein FII, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-05 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XPM
 
 | | Mature virion portal of phage lambda with DNA | | Descriptor: | DNA (104-MER), DNA (92-MER), Head completion protein, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-04 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XOT
 
 | | Prohead portal of bacteriophage lambda | | Descriptor: | Portal protein B | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8XOW
 
 | | Mature virion portal of bacteriophage lambda | | Descriptor: | Head completion protein, Head-tail connector protein FII, Portal protein B, ... | | Authors: | Wang, J.W, Gu, Z.W. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structural morphing in the viral portal vertex of bacteriophage lambda.
J.Virol., 98, 2024
|
|
8HVI
 
 | | Activation mechanism of GPR132 by compound NOX-6-7 | | Descriptor: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | Deposit date: | 2022-12-26 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
3NTG
 
 | | Crystal structure of COX-2 with selective compound 23d-(R) | | Descriptor: | (2R)-6,8-dichloro-7-(2-methylpropoxy)-2-(trifluoromethyl)-2H-chromene-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J.L, Limburg, D, Graneto, M.J, Carter, J.C, Talley, J.J, Kiefer, J.R. | | Deposit date: | 2010-07-04 | | Release date: | 2010-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The novel benzopyran class of selective cyclooxygenase-2 inhibitors. Part 2: The second clinical candidate having a shorter and favorable human half-life.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5Y0A
 
 | |
8IGA
 
 | |
7XVI
 
 | |
7XVK
 
 | |
7MKK
 
 | |
6LP3
 
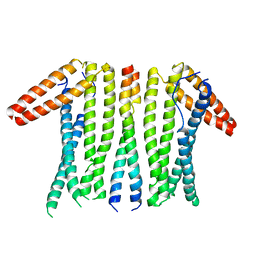 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
7XZ9
 
 | |
7XZD
 
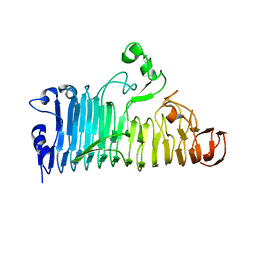 | |
7XZ8
 
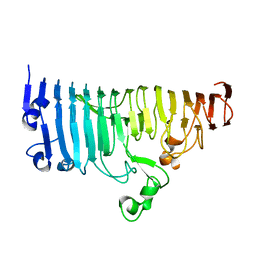 | |
7XZB
 
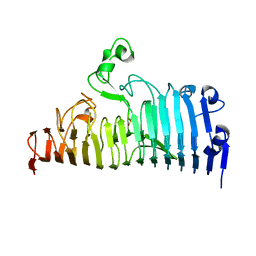 | |
7XZA
 
 | |
7XZE
 
 | |
7XZ7
 
 | |
7XZC
 
 | |
6J5S
 
 | | Crystal structure of human HINT1 mutant complexing with AP5A | | Descriptor: | BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ETHANESULFONIC ACID, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-11 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J53
 
 | | Crystal structure of human HINT1 complexing with ATP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J58
 
 | | Crystal structure of human HINT1 complexing with AP4A | | Descriptor: | ADENOSINE MONOPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-10 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
6J64
 
 | | Crystal structure of human HINT1 mutant complexing with AP4A | | Descriptor: | 2-AMINOETHANESULFONIC ACID, BIS(ADENOSINE)-5'-TETRAPHOSPHATE, Histidine triad nucleotide-binding protein 1 | | Authors: | Wang, J, Fang, P, Guo, M. | | Deposit date: | 2019-01-14 | | Release date: | 2019-09-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Second messenger Ap4A polymerizes target protein HINT1 to transduce signals in Fc epsilon RI-activated mast cells.
Nat Commun, 10, 2019
|
|
