6M6F
 
 | |
8GHS
 
 | | Empty HBV Cp183 capsid with importin-beta, subparticle reconstruction at 2-fold location | | Descriptor: | Capsid protein | | Authors: | Kim, C, Schlicksup, C.J, Hadden-Perilla, J.A, Wang, J.C.-Y, Zlotnick, A. | | Deposit date: | 2023-03-10 | | Release date: | 2023-08-09 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the Hepatitis B virus capsid quasi-6-fold with a trapped C-terminal domain reveals capsid movements associated with domain exit.
J.Biol.Chem., 299, 2023
|
|
5JPS
 
 | |
3KD7
 
 | |
3KHF
 
 | | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Microtubule-associated serine/threonine-protein kinase 3 | | Authors: | Roos, A, Elkins, J, Savitsky, P, Wang, J, Ugochukwu, E, Murray, J, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of the PDZ domain of human Microtubule Associated Serine/Threonine Kinase 3 (MAST3)
To be Published
|
|
6WGB
 
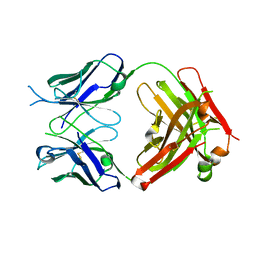 | | Crystal structure of the fab portion of dupilumab | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
6WGL
 
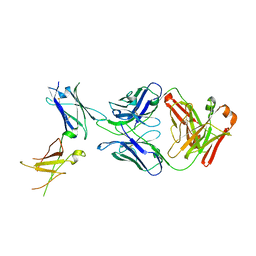 | | Dupilumab fab with Crystal Kappa design complexed with human IL-4 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Dupilumab Fab heavy chain, ... | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
4RHR
 
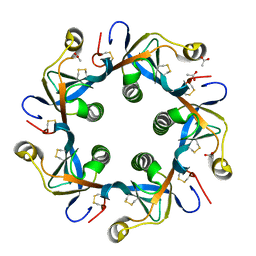 | | Crystal structure of PltB | | Descriptor: | ACETATE ION, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2014-12-17 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
4RHS
 
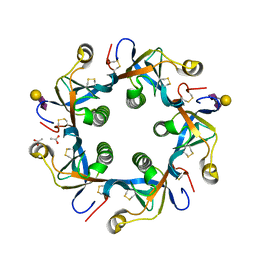 | | Crystal structure of GD2 bound PltB | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid-(2-8)-N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, Putative pertussis-like toxin subunit | | Authors: | Gao, X, Wang, J, Galan, J. | | Deposit date: | 2014-10-02 | | Release date: | 2014-10-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9211 Å) | | Cite: | Host adaptation of a bacterial toxin from the human pathogen salmonella typhi.
Cell(Cambridge,Mass.), 159, 2014
|
|
6IFN
 
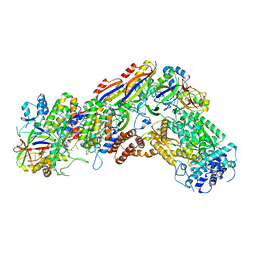 | | Crystal structure of Type III-A CRISPR Csm complex | | Descriptor: | MANGANESE (II) ION, RNA (32-MER), Type III-A CRISPR-associated RAMP protein Csm3, ... | | Authors: | You, L, Wang, J, Wang, Y. | | Deposit date: | 2018-09-20 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure Studies of the CRISPR-Csm Complex Reveal Mechanism of Co-transcriptional Interference
Cell, 176, 2019
|
|
6WG8
 
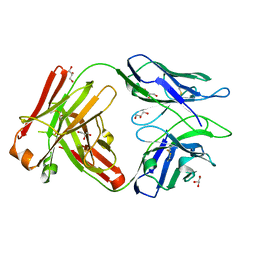 | | Fab portion of dupilumab with Crystal Kappa design | | Descriptor: | Dupilumab Fab heavy chain, Dupilumab Fab light chain, GLYCEROL | | Authors: | Druzina, Z, Atwell, S, Pustilnik, A, Antonysamy, S, Ho, C, Lieu, R, Hendle, J, Benach, J, Wang, J. | | Deposit date: | 2020-04-05 | | Release date: | 2020-09-16 | | Last modified: | 2020-09-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Rapid and robust antibody Fab fragment crystallization utilizing edge-to-edge beta-sheet packing.
Plos One, 15, 2020
|
|
3J32
 
 | | An asymmetric unit map from electron cryo-microscopy of Haliotis diversicolor molluscan hemocyanin isoform 1 (HdH1) | | Descriptor: | Hemocyanin isoform 1 | | Authors: | Zhang, Q, Dai, X, Cong, Y, Zhang, J, Chen, D.-H, Dougherty, M, Wang, J, Ludtke, S, Schmid, M.F, Chiu, W. | | Deposit date: | 2013-02-20 | | Release date: | 2013-04-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of a molluscan hemocyanin suggests its allosteric mechanism.
Structure, 21, 2013
|
|
4RVZ
 
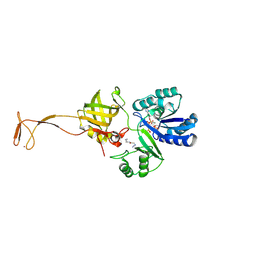 | | Crystal structure of tRNA fluorescent labeling enzyme | | Descriptor: | MAGNESIUM ION, N-(4-aminobutyl)-2-azidoacetamide, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Dong, J, Li, F, Wang, J, Gong, W. | | Deposit date: | 2014-11-29 | | Release date: | 2015-03-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A covalent approach for site-specific RNA labeling in Mammalian cells.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1ZHA
 
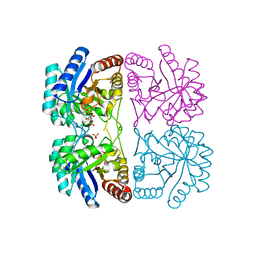 | | A. aeolicus KDO8PS R106G mutant in complex with PEP and R5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop.
Biochemistry, 44, 2005
|
|
2Z3B
 
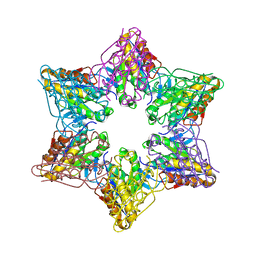 | | Crystal Structure of Bacillus Subtilis CodW, a non-canonical HslV-like peptidase with an impaired catalytic apparatus | | Descriptor: | ATP-dependent protease hslV, SODIUM ION | | Authors: | Rho, S.H, Park, H.H, Kang, G.B, Lim, Y.J, Kang, M.S, Lim, B.K, Seong, I.S, Chung, C.H, Wang, J, Eom, S.H. | | Deposit date: | 2007-06-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Bacillus subtilis CodW, a noncanonical HslV-like peptidase with an impaired catalytic apparatus
Proteins, 71, 2007
|
|
6XBI
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW248 | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-06 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
6KHX
 
 | | Crystal structure of Prx from Akkermansia muciniphila | | Descriptor: | CALCIUM ION, Peroxiredoxin | | Authors: | Li, M, Wang, J, Xu, W, Wang, Y, Zhang, M, Wang, M. | | Deposit date: | 2019-07-16 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of Akkermansia muciniphila peroxiredoxin reveals a novel regulatory mechanism of typical 2-Cys Prxs by a distinct loop.
Febs Lett., 594, 2020
|
|
6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
1ZJI
 
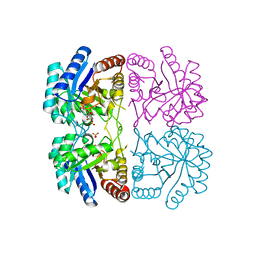 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
8XOX
 
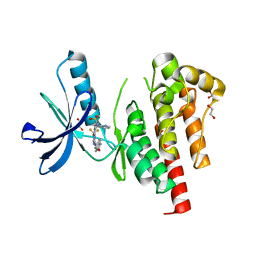 | | The Crystal Structure of FAK2 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide, Protein-tyrosine kinase 2-beta | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-01-02 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of FAK2 from Biortus.
To Be Published
|
|
8ZH5
 
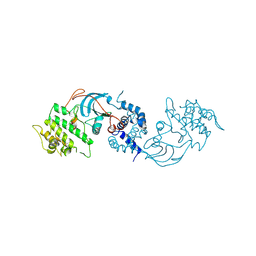 | |
8YHP
 
 | | Structure of the PGK1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, Phosphoglycerate kinase 1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Wang, J. | | Deposit date: | 2024-02-28 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the PGK1 from Biortus.
To Be Published
|
|
4Q0F
 
 | | Crystal Structure of Thermotoga maritima FtsH Periplasmic domain | | Descriptor: | ATP-dependent zinc metalloprotease FtsH | | Authors: | An, J.Y, Sharif, H, Barrera, F.N, Karabadzhak, A, Kang, G.B, Park, K.J, Sakkiah, S, Lee, K.W, Lee, S, Engelman, D.M, Wang, J, Eom, S.H. | | Deposit date: | 2014-04-01 | | Release date: | 2015-04-01 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structures of periplasmic and transmembrane domains of FtsH suggest a reverse translocon mechanism for protein extraction from membrane
To be Published
|
|
