1MIW
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
5UFL
 
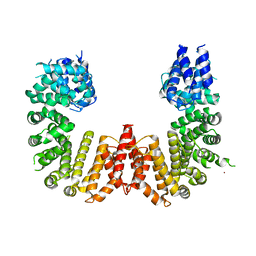 | | Crystal structure of a CIP2A core domain | | Descriptor: | Protein CIP2A, ZINC ION | | Authors: | Wang, Z, Wang, J, Rao, Z, Xu, W. | | Deposit date: | 2017-01-04 | | Release date: | 2017-02-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Oncoprotein CIP2A is stabilized via interaction with tumor suppressor PP2A/B56.
EMBO Rep., 18, 2017
|
|
1W63
 
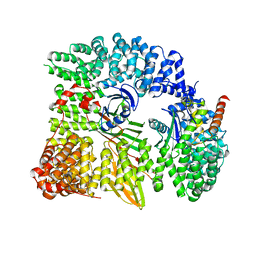 | | AP1 clathrin adaptor core | | Descriptor: | ADAPTER-RELATED PROTEIN COMPLEX 1 BETA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 GAMMA 1 SUBUNIT, ADAPTER-RELATED PROTEIN COMPLEX 1 SIGMA 1A SUBUNIT, ... | | Authors: | Heldwein, E, Macia, E, Wang, J, Yin, H.L, Kirchhausen, T, Harrison, S.C. | | Deposit date: | 2004-08-12 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure of the Clathrin Adaptor Protein 1 Core
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1TF7
 
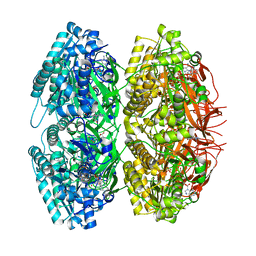 | | Crystal Structure of Circadian Clock Protein KaiC | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC | | Authors: | Pattanayek, R, Wang, J, Mori, T, Xu, Y, Johnson, C.H, Egli, M. | | Deposit date: | 2004-05-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Visualizing a Circadian Clock Protein; Crystal Structure of KaiC and Functional Insights
Mol.Cell, 15, 2004
|
|
6WB8
 
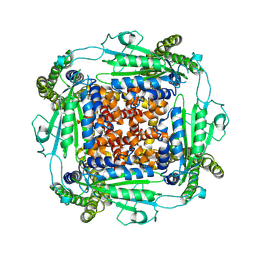 | | Cryo-EM structure of PKD2 C331S disease variant | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystin-2 | | Authors: | Cao, E, Wang, J, Decaen, P.G. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular dysregulation of ciliary polycystin-2 channels caused by variants in the TOP domain.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1MIY
 
 | | Crystal structure of Bacillus stearothermophilus CCA-adding enzyme in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, tRNA CCA-adding enzyme | | Authors: | Li, F, Xiong, Y, Wang, J, Cho, H.D, Weiner, A.M, Steitz, T.A. | | Deposit date: | 2002-08-23 | | Release date: | 2002-12-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and
its complexes with ATP or CTP
Cell(Cambridge,Mass.), 111, 2002
|
|
6VZP
 
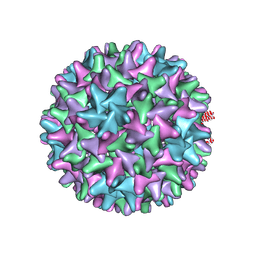 | | HBV wild type capsid | | Descriptor: | Capsid protein | | Authors: | Zhao, Z, Wang, J, Zlotnick, A. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-30 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | The Integrity of the Intradimer Interface of the Hepatitis B Virus Capsid Protein Dimer Regulates Capsid Self-Assembly.
Acs Chem.Biol., 15, 2020
|
|
1M60
 
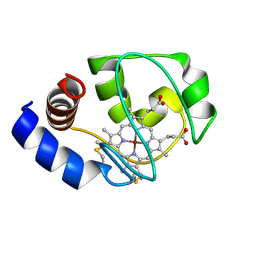 | | Solution Structure of Zinc-substituted cytochrome c | | Descriptor: | ZINC SUBSTITUTED HEME C, Zinc-substituted cytochrome c | | Authors: | Qian, C, Yao, Y, Tong, Y, Wang, J, Tang, W. | | Deposit date: | 2002-07-11 | | Release date: | 2002-08-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of zinc-substituted cytochrome c.
J.Biol.Inorg.Chem., 8, 2003
|
|
1NGM
 
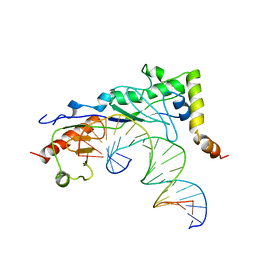 | | Crystal structure of a yeast Brf1-TBP-DNA ternary complex | | Descriptor: | 5'-D(*AP*AP*AP*AP*AP*AP*CP*AP*TP*TP*TP*TP*TP*TP*TP*AP*TP*AP*G)-3', 5'-D(*CP*TP*AP*TP*AP*AP*AP*AP*AP*AP*AP*TP*GP*TP*TP*TP*TP*TP*T)-3', Transcription factor IIIB BRF1 subunit, ... | | Authors: | Juo, Z.S, Kassavetis, G.A, Wang, J, Geiduschek, E.P, Sigler, P.B. | | Deposit date: | 2002-12-17 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal structure of a transcription factor IIIB core interface ternary complex
Nature, 422, 2003
|
|
5V5M
 
 | |
5V5L
 
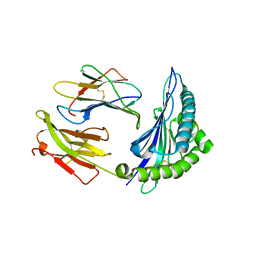 | |
1SZT
 
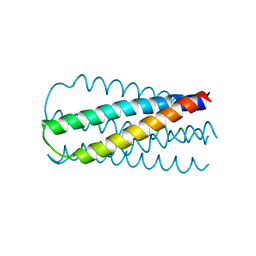 | |
1TYE
 
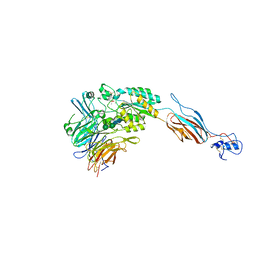 | | Structural basis for allostery in integrins and binding of ligand-mimetic therapeutics to the platelet receptor for fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CACODYLATE ION, CALCIUM ION, ... | | Authors: | Xiao, T, Takagi, J, Coller, B.S, Wang, J.-H, Springer, T.A. | | Deposit date: | 2004-07-07 | | Release date: | 2004-10-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics
Nature, 432, 2004
|
|
6ULW
 
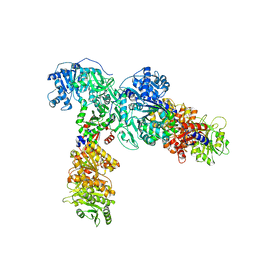 | | Adenylation, ketoreductase, and pseudo Asub multidomain structure of a keto acid-selecting NRPS module | | Descriptor: | Amino acid adenylation domain-containing protein, CALCIUM ION, MAGNESIUM ION | | Authors: | Alonzo, D.A, Wang, J, Chiche-Lapierre, C, Schmeing, T.M. | | Deposit date: | 2019-10-08 | | Release date: | 2020-02-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of keto acid utilization in nonribosomal depsipeptide synthesis.
Nat.Chem.Biol., 16, 2020
|
|
1LS4
 
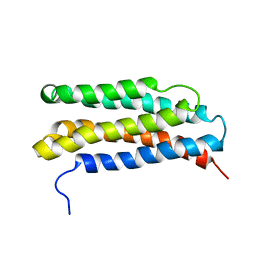 | |
1T0P
 
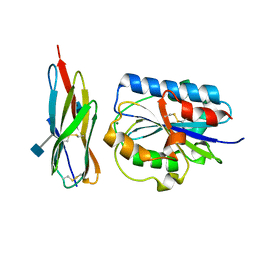 | | Structural Basis of ICAM recognition by integrin alpahLbeta2 revealed in the complex structure of binding domains of ICAM-3 and alphaLbeta2 at 1.65 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, Intercellular adhesion molecule-3, ... | | Authors: | Song, G, Yang, Y.T, Liu, J.H, Shimaoko, M, Springer, T.A, Wang, J.H. | | Deposit date: | 2004-04-12 | | Release date: | 2005-03-08 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | An atomic resolution view of ICAM recognition in a complex between the binding domains of ICAM-3 and integrin alphaLbeta2.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1MQA
 
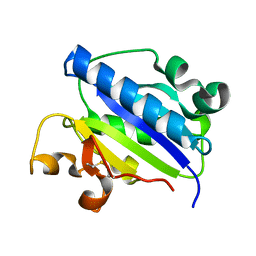 | | Crystal structure of high affinity alphaL I domain in the absence of ligand or metal | | Descriptor: | Integrin alpha-L | | Authors: | Shimaoka, T, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, Zhang, R, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MQ9
 
 | | Crystal structure of high affinity alphaL I domain with ligand mimetic crystal contact | | Descriptor: | Integrin alpha-L, MANGANESE (II) ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1SJW
 
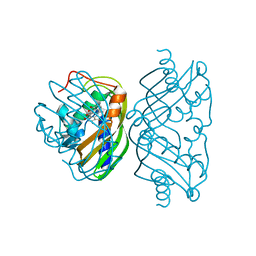 | | Structure of polyketide cyclase SnoaL | | Descriptor: | METHYL 5,7-DIHYDROXY-2-METHYL-4,6,11-TRIOXO-3,4,6,11-TETRAHYDROTETRACENE-1-CARBOXYLATE, nogalonic acid methyl ester cyclase | | Authors: | Sultana, A, Kallio, P, Jansson, A, Wang, J.S, Neimi, J, Mantsala, P, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-03-04 | | Release date: | 2004-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the polyketide cyclase SnoaL reveals a novel mechanism for enzymatic aldol condensation.
Embo J., 23, 2004
|
|
6A7H
 
 | | Bacterial protein toxins | | Descriptor: | RTX toxin, SULFATE ION | | Authors: | Kim, M.H, Hwang, J, Jang, S.Y. | | Deposit date: | 2018-07-03 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural basis of inactivation of Ras and Rap1 small GTPases by Ras/Rap1-specific endopeptidase from the sepsis-causing pathogenVibrio vulnificus
J. Biol. Chem., 293, 2018
|
|
1L6Z
 
 | | CRYSTAL STRUCTURE OF MURINE CEACAM1A[1,4]: A CORONAVIRUS RECEPTOR AND CELL ADHESION MOLECULE IN THE CEA FAMILY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, biliary glycoprotein C | | Authors: | Tan, K, Zelus, B.D, Meijers, R, Liu, J.-H, Bergelson, J.M, Duke, N, Zhang, R, Joachimiak, A, Holmes, K.V, Wang, J.-H. | | Deposit date: | 2002-03-14 | | Release date: | 2002-09-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | CRYSTAL STRUCTURE OF MURINE sCEACAM1a[1,4]: A CORONAVIRUS RECEPTOR IN THE CEA FAMILY
Embo J., 21, 2002
|
|
1MQ8
 
 | | Crystal structure of alphaL I domain in complex with ICAM-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Integrin alpha-L, ... | | Authors: | Shimaoka, M, Xiao, T, Liu, J.-H, Yang, Y, Dong, Y, Jun, C.-D, McCormack, A, Zhang, R, Joachimiak, A, Takagi, J, Wang, J.-H, Springer, T.A. | | Deposit date: | 2002-09-15 | | Release date: | 2003-01-14 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the aL I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
1MJN
 
 | | Crystal Structure of the intermediate affinity aL I domain mutant | | Descriptor: | Integrin alpha-L, MAGNESIUM ION | | Authors: | Shimaoka, M, Xiao, T, Liu, J.H, Yang, Y.T, Dong, Y.C, Jun, C.D, McCormack, A, Zhang, R.G, Wang, J.H, Springer, T.A. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-28 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the alphaL I Domain and its Complex with ICAM-1 reveal a Shape-shifting Pathway for Integrin Regulation
Cell(Cambridge,Mass.), 112, 2003
|
|
8D8O
 
 | | Cryo-EM structure of substrate unbound PAPP-A | | Descriptor: | Pappalysin-1, ZINC ION | | Authors: | Judge, R.A, Jain, R, Hao, Q, Ouch, C, Sridar, J, Smith, C.L, Wang, J.C.K, Eaton, D. | | Deposit date: | 2022-06-08 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of the PAPP-ABP5 complex reveals mechanism of substrate recognition
Nat Commun, 13, 2022
|
|
1K1D
 
 | | Crystal structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Cheon, Y.H, Kim, H.S, Han, K.H, Abendroth, J, Niefind, K, Schomburg, D, Wang, J, Kim, Y. | | Deposit date: | 2001-09-25 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity.
Biochemistry, 41, 2002
|
|
