8Y8E
 
 | | Structure of HCoV-HKU1C spike in the inactive-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y87
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-1up conformation with 1TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y89
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Wang, H.F, Zhang, X, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8A
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8D
 
 | | Structure of HCoV-HKU1C spike in the inactive-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8F
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8C
 
 | | Structure of HCoV-HKU1C spike in the inactive-closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y88
 
 | | Structure of HCoV-HKU1C spike in the functionally anchored-2up conformation with 2TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8H
 
 | | Structure of HCoV-HKU1C spike in the glycan-activated-2up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8J
 
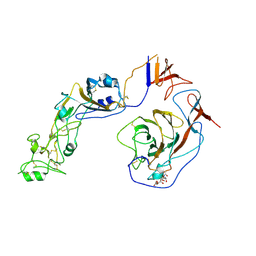 | | Local structure of HCoV-HKU1C spike in complex with glycan | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y8G
 
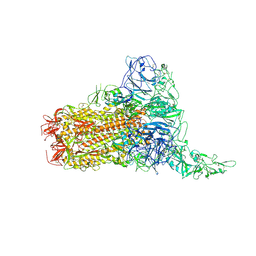 | | Structure of HCoV-HKU1C spike in the glycan-activated-1up conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-06 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
8Y7X
 
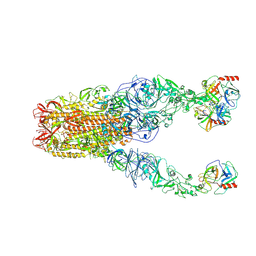 | | Structure of HCoV-HKU1A spike in the functionally anchored-3up conformation with 3TMPRSS2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Lu, Y.C, Zhang, X, Wang, H.F, Liu, X.C, Sun, L, Yang, H.T. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | TMPRSS2 and glycan receptors synergistically facilitate coronavirus entry.
Cell, 187, 2024
|
|
5KQA
 
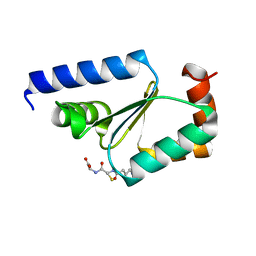 | | Crystal structure of buckwheat glutaredoxin-glutathione complex | | Descriptor: | GLUTATHIONE, Glutaredoxin-glutathione complex | | Authors: | Zhang, X, Wang, W, Zhao, Y, Wang, Z, Wang, H. | | Deposit date: | 2016-07-06 | | Release date: | 2017-07-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural insights into the binding of buckwheat glutaredoxin with GSH and regulation of its catalytic activity
J. Inorg. Biochem., 173, 2017
|
|
4AOJ
 
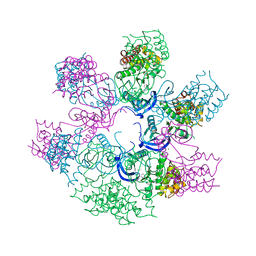 | | Human TrkA in complex with the inhibitor AZ-23 | | Descriptor: | 5-chloranyl-N2-[(1S)-1-(5-fluoranylpyridin-2-yl)ethyl]-N4-(3-propan-2-yloxy-1H-pyrazol-5-yl)pyrimidine-2,4-diamine, HIGH AFFINITY NERVE GROWTH FACTOR RECEPTOR, ZINC ION | | Authors: | Wang, T, Lamb, M.L, Block, M.H, Davies, A.M, Han, Y, Hoffmann, E, Ioannidis, S, Josey, J.A, Liu, Z, Lyne, P.D, MacIntyre, T, Mohr, P.J, Omer, C.A, Sjogren, T, Thress, K, Wang, B, Wang, H, Yu, D, Zhang, H. | | Deposit date: | 2012-03-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Discovery of Disubstituted Imidazo[4,5-B]Pyridines and Purines as Potent Trka Inhibitors
Acs Med.Chem.Lett., 3, 2012
|
|
5WH6
 
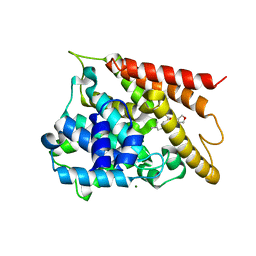 | | Crystal structure of PDE4D2 in complex with inhibitor (S_Zl-n-91) | | Descriptor: | 1-[4-(difluoromethoxy)-3-{[(3S)-oxolan-3-yl]oxy}phenyl]-3-methylbutan-1-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ke, H, Wang, H. | | Deposit date: | 2017-07-14 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification of a PDE4-Specific Pocket for the Design of Selective Inhibitors.
Biochemistry, 57, 2018
|
|
4V8X
 
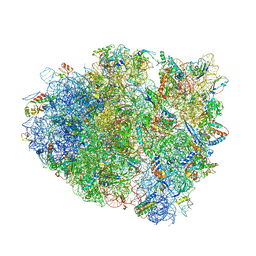 | | Structure of Thermus thermophilus ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Feng, S, Chen, Y, Kamada, K, Wang, H, Tang, K, Wang, M, Gao, Y.G. | | Deposit date: | 2013-07-19 | | Release date: | 2014-07-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Yoeb-Ribosome Structure: A Canonical Rnase that Requires the Ribosome for its Specific Activity.
Nucleic Acids Res., 41, 2013
|
|
6M1H
 
 | | CryoEM structure of human PAC1 receptor in complex with maxadilan | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Song, X, Wang, J, Zhang, D, Wang, H.W, Ma, Y. | | Deposit date: | 2020-02-26 | | Release date: | 2020-03-11 | | Last modified: | 2020-05-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism.
Cell Res., 30, 2020
|
|
6L82
 
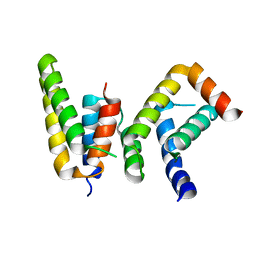 | | Crystal structure of Chaetomium GCP5 N-terminus and Mozart1 | | Descriptor: | Mozart1, Spindle pole body component | | Authors: | Huang, T.L, Wang, H.J, Wang, S.W, Hsia, K.C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-07-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.24103618 Å) | | Cite: | Promiscuous Binding of Microprotein Mozart1 to gamma-Tubulin Complex Mediates Specific Subcellular Targeting to Control Microtubule Array Formation.
Cell Rep, 31, 2020
|
|
6LXD
 
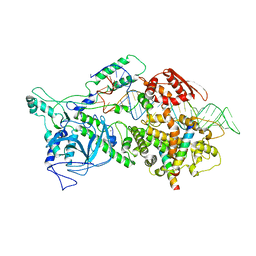 | | Pri-miRNA bound DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
6LXE
 
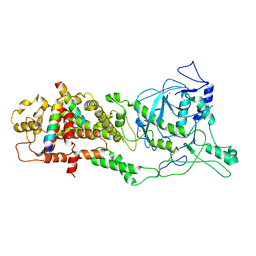 | | DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
4RA2
 
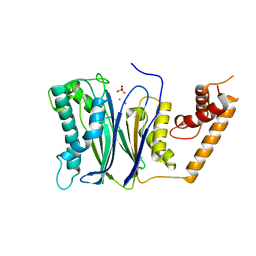 | | PP2Ca | | Descriptor: | MANGANESE (II) ION, PHOSPHATE ION, Protein phosphatase 1A | | Authors: | Pan, C, Tang, J.Y, Xu, Y.F, Xiao, P, Liu, H.D, Wang, H.A, Wang, W.B, Meng, F.G, Yu, X, Sun, J.P. | | Deposit date: | 2014-09-09 | | Release date: | 2015-05-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The catalytic role of the M2 metal ion in PP2Ca
SCI REP, 2015
|
|
5X1G
 
 | | WHAMM's Microtubule binding motif | | Descriptor: | WASP homolog-associated protein with actin, membranes and microtubules | | Authors: | Liu, T, Wang, H.W. | | Deposit date: | 2017-01-25 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Insights of WHAMM's Interaction with Microtubules by Cryo-EM
J. Mol. Biol., 429, 2017
|
|
4ZNQ
 
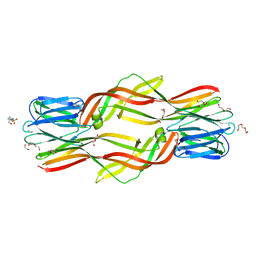 | | Crystal structure of Dln1 complexed with Man(alpha1-2)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNO
 
 | | Crystal structure of Dln1 complexed with sucrose | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, Natterin-like protein, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
4ZNR
 
 | | Crystal structure of Dln1 complexed with Man(alpha1-3)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
