8T91
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-IP6 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T95
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-(1,2,4,5)-IP4 | | Descriptor: | (1S,3R,4R,6S)-1,3,4,6-TETRAPKISPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8T8Z
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and myo-(1OH)IP5 | | Descriptor: | (1S,2R,3S,4S,5S,6S)-6-hydroxycyclohexane-1,2,3,4,5-pentayl pentakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
5G2X
 
 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | 5'-R(*CP*AP*CP*AP*UP*CP*CP*AP*UP*AP*AP*CP)-3', GROUP II INTRON, GROUP II INTRON-ENCODED PROTEIN LTRA | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-11 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5G2Y
 
 | | Structure a of Group II Intron Complexed with its Reverse Transcriptase | | Descriptor: | GROUP II INTRON | | Authors: | Qu, G, Kaushal, P.S, Wang, J, Shigematsu, H, Piazza, C.L, Agrawal, R.K, Belfort, M, Wang, H.W. | | Deposit date: | 2016-04-16 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of a Group II Intron in Complex with its Reverse Transcriptase.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5G06
 
 | | Cryo-EM structure of yeast cytoplasmic exosome | | Descriptor: | EXOSOME COMPLEX COMPONENT CSL4, EXOSOME COMPLEX COMPONENT MTR3, EXOSOME COMPLEX COMPONENT RRP4, ... | | Authors: | Liu, J.J, Niu, C.Y, Wu, Y, Tan, D, Wang, Y, Ye, M.D, Liu, Y, Zhao, W.W, Zhou, K, Liu, Q.S, Dai, J.B, Yang, X.R, Dong, M.Q, Huang, N, Wang, H.W. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-15 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryoem Structure of Yeast Cytoplasmic Exosome Complex.
Cell Res., 26, 2016
|
|
6QIM
 
 | | Structure of AtPIP2;4 | | Descriptor: | Probable aquaporin PIP2-4 | | Authors: | Schoebel, S, Wang, H. | | Deposit date: | 2019-01-21 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Characterization of aquaporin-driven hydrogen peroxide transport.
Biochim Biophys Acta Biomembr, 1862, 2020
|
|
4JPZ
 
 | | Voltage-gated sodium channel 1.2 C-terminal domain in complex with FGF13U and Ca2+/calmodulin | | Descriptor: | CALCIUM ION, Calmodulin, Fibroblast growth factor 13, ... | | Authors: | Wang, C, Chung, B.C, Yan, H, Wang, H.G, Lee, S.Y, Pitt, G.S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural analyses of Ca(2+)/CaM interaction with NaV channel C-termini reveal mechanisms of calcium-dependent regulation.
Nat Commun, 5, 2014
|
|
2FH3
 
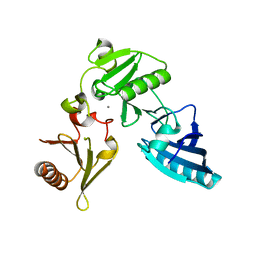 | | C-terminal half of gelsolin soaked in low calcium at pH 8 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
2FH2
 
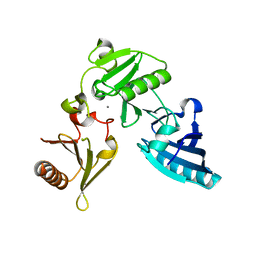 | | C-terminal half of gelsolin soaked in EGTA at pH 4.5 | | Descriptor: | CALCIUM ION, Gelsolin | | Authors: | Chumnarnsilpa, S, Loonchanta, A, Xue, B, Choe, H, Urosev, D, Wang, H, Burtnick, L.D, Robinson, R.C. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Calcium ion exchange in crystalline gelsolin
J.Mol.Biol., 357, 2006
|
|
4C60
 
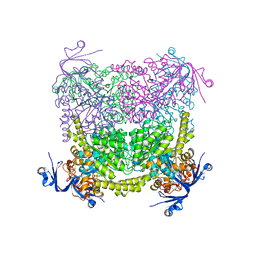 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
4C5Y
 
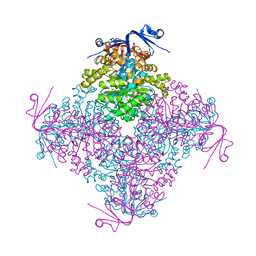 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE, ZINC ION | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
8T8M
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nb43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-22 | | Release date: | 2023-10-11 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T6J
 
 | | CDPPB-bound inactive mGlu5 | | Descriptor: | 3-cyano-N-(1,3-diphenyl-1H-pyrazol-5-yl)benzamide, Metabotropic glutamate receptor 5 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-16 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
8T7H
 
 | | Quis-bound intermediate mGlu5 | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Metabotropic glutamate receptor 5, Nanobody 43 | | Authors: | Krishna Kumar, K, Wang, H, Kobilka, B.K. | | Deposit date: | 2023-06-20 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Stepwise activation of a metabotropic glutamate receptor.
Nature, 629, 2024
|
|
4C65
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
8W7T
 
 | | Fe-O nanocluster of form-VII in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W73
 
 | | Fe-O nanocluster of form-I in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7V
 
 | | Udif-E164A-E168A soaking in Fe2+ solution for 50 minutes | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W79
 
 | | Fe-O nanocluster of form-III in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W6Y
 
 | | Ferritin from Ureaplasma diversum soaking in Fe2+ solution for 10 min | | Descriptor: | FE (III) ION, Ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7O
 
 | | Fe-O nanocluster of form-V in the 4-fold channel of Ureaplasma diversum ferritin | | Descriptor: | FE (III) ION, Ferritin, MAGNESIUM ION | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W6M
 
 | | Native strucutre of ferritin from Ureaplasma diversum | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W7U
 
 | | Mutant of ferritin from Ureaplasma diversum (Udif-E164A-E168A) without soaking | | Descriptor: | FE (III) ION, Ferritin, MAGNESIUM ION | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-31 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8W6S
 
 | | Ferritin from Ureaplasma diversum soaking in Fe2+ solution for 2 min | | Descriptor: | CHLORIDE ION, FE (III) ION, Ferritin, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-08-29 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
