8PM3
 
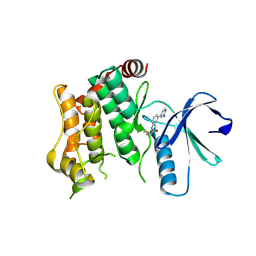 | | Crystal structure of MAP2K6 with a covalent compound GCL94 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, ~{N}-[3-(2-azanylpyridin-4-yl)phenyl]propanamide | | Authors: | Wang, G.Q, Seidler, N, Roehm, S, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-06-28 | | Release date: | 2023-09-20 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
8P7J
 
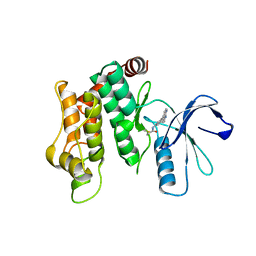 | | Crystal structure of MAP2K6 with a covalent compound GCL96 | | Descriptor: | Dual specificity mitogen-activated protein kinase kinase 6, N-[3-(1H-pyrrolo[2,3-b]pyridin-4-yl)phenyl]prop-2-enamide | | Authors: | Wang, G.Q, Seidler, N, Roehm, S, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-30 | | Release date: | 2023-07-05 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8ZJW
 
 | | Cryo-EM structure of photosynthetic LH1' complex of Roseospirillum parvum | | Descriptor: | Alpha subunit of light-harvesting 1 complex, BACTERIOCHLOROPHYLL A, Beta subunit of light-harvesting 1 complex, ... | | Authors: | Wang, G.-L, Wang, X.-P, Yu, L.-J. | | Deposit date: | 2024-05-15 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Insights into the divergence of the photosynthetic LH1 complex obtained from structural analysis of the unusual photocomplexes of Roseospirillum parvum.
Commun Biol, 7, 2024
|
|
8ZK2
 
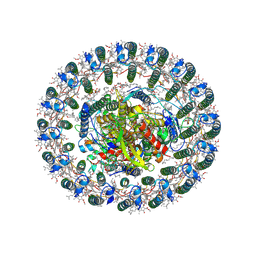 | | Cryo-EM structure of photosynthetic LH1-RC core complex of Roseospirillum parvum | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Alpha subunit of light-harvesting 1 complex, BACTERIOCHLOROPHYLL A, ... | | Authors: | Wang, G.-L, Wang, X.-P, Yu, L.-J. | | Deposit date: | 2024-05-15 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Insights into the divergence of the photosynthetic LH1 complex obtained from structural analysis of the unusual photocomplexes of Roseospirillum parvum.
Commun Biol, 7, 2024
|
|
6NM3
 
 | | NMR structure of WW295 | | Descriptor: | WW295 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Two distinct amphipathic peptide antibiotics with systemic efficacy.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6NM2
 
 | | NMR Structure of WW291 | | Descriptor: | WW291 peptide | | Authors: | Wang, G, Zarena, D. | | Deposit date: | 2019-01-10 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | The pi Configuration of the WWW Motif of a Short Trp-Rich Peptide Is Critical for Targeting Bacterial Membranes, Disrupting Preformed Biofilms, and Killing Methicillin-Resistant Staphylococcus aureus.
Biochemistry, 56, 2017
|
|
9F81
 
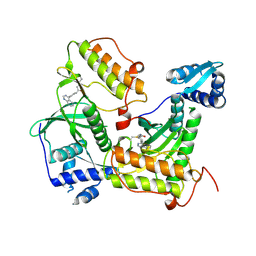 | | Crystal structure of RIOK2 with a covalent compound GCL 47 | | Descriptor: | N-(4-quinazolin-4-ylphenyl)propanamide, Serine/threonine-protein kinase RIO2 | | Authors: | Wang, G.Q, Wydra, V, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-05-06 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1XOG
 
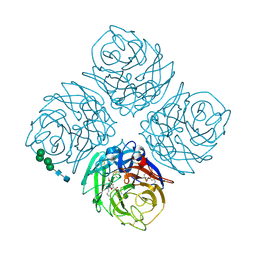 | | N9 Tern Influenza neuraminidase complexed with a 2,5-Disubstituted tetrahydrofuran-5-carboxylic acid | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-2,5-ANHYDRO-3,4-DIDEOXY-4-(METHOXYCARBONYL)PENTONIC ACID, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
9F31
 
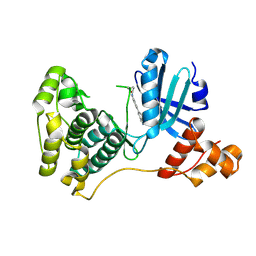 | | Crystal structure of MELK with a covalent compound GCL 99 | | Descriptor: | Maternal embryonic leucine zipper kinase, N-[4-(1H-indazol-5-yl)phenyl]propanamide | | Authors: | Wang, G.Q, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-24 | | Release date: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
9F32
 
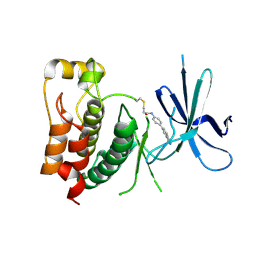 | | Crystal structure of ULK1 with a covalent compound GCL 99 | | Descriptor: | N-[4-(1H-indazol-5-yl)phenyl]propanamide, Serine/threonine-protein kinase ULK1 | | Authors: | Wang, G.Q, Seidler, N, Gehringer, M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-24 | | Release date: | 2025-01-08 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing the Protein Kinases' Cysteinome by Covalent Fragments.
Angew.Chem.Int.Ed.Engl., 64, 2025
|
|
1XOE
 
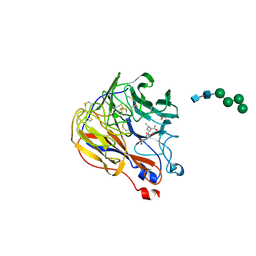 | | N9 Tern influenza neuraminidase complexed with (2R,4R,5R)-5-(1-Acetylamino-3-methyl-butyl-pyrrolidine-2, 4-dicarobyxylic acid 4-methyl esterdase complexed with | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[1-(ACETYLAMINO)-3-METHYLBUTYL]-4-(METHOXYCARBONYL)PROLINE, Neuraminidase, ... | | Authors: | Wang, G.T, Wang, S, Gentles, R, Sowin, T, Maring, C.J, Kempf, D.J, Kati, W.M, Stoll, V, Stewart, K.D, Laver, G. | | Deposit date: | 2004-10-06 | | Release date: | 2005-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and structural analysis of inhibitors of influenza neuraminidase
containing a 2,3-disubstituted tetrahydrofuran-5-carboxylic acid core.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1VM5
 
 | |
1VM2
 
 | |
1VM4
 
 | |
1VM3
 
 | |
8Q1Z
 
 | |
6XSP
 
 | |
6XSQ
 
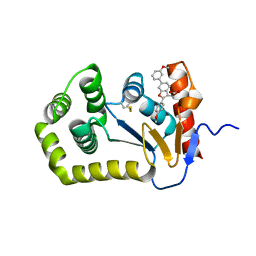 | |
6XT3
 
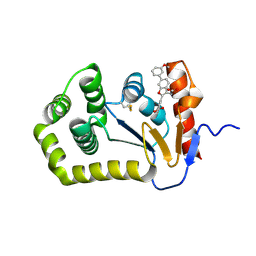 | |
6PVY
 
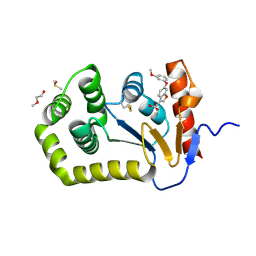 | |
6PVZ
 
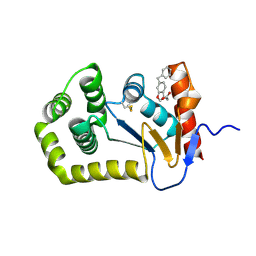 | |
1NH9
 
 | | Crystal Structure of a DNA Binding Protein Mja10b from the hyperthermophile Methanococcus jannaschii | | Descriptor: | DNA-binding protein Alba | | Authors: | Wang, G, Bartlam, M, Guo, R, Yang, H, Xue, H, Liu, Y, Huang, L, Rao, Z. | | Deposit date: | 2002-12-19 | | Release date: | 2003-12-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a DNA binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii
Protein Sci., 12, 2003
|
|
9FR2
 
 | |
9FET
 
 | |
