6WL9
 
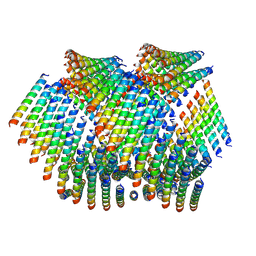 | | Cryo-EM of Form 2 like peptide filament, Form2a | | Descriptor: | peptide Form2a | | Authors: | Wang, F, Beltran, L.C, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
5D6Y
 
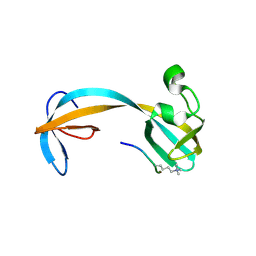 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A complexed with histone H3K23me3 | | Descriptor: | Lysine-specific demethylase 4A, peptide H3K23me3 (19-28) | | Authors: | Wang, F, Su, Z, Miller, M.D, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2016-02-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.287 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
6UHY
 
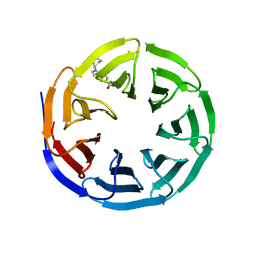 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzimidazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.W. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
6UHZ
 
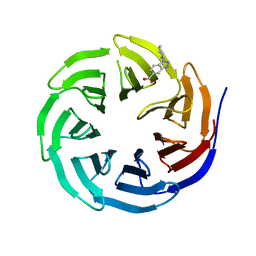 | | WDR5 in complex with Myc site fragment inhibitor | | Descriptor: | 1-cyclohexyl-1H-benzotriazole-5-carboxylic acid, WDR5 | | Authors: | Wang, F, Fesik, S.w. | | Deposit date: | 2019-09-29 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.258 Å) | | Cite: | Discovery of WD Repeat-Containing Protein 5 (WDR5)-MYC Inhibitors Using Fragment-Based Methods and Structure-Based Design.
J.Med.Chem., 63, 2020
|
|
8ZH5
 
 | |
6W8U
 
 | | Cryo-EM of the Pyrobaculum arsenaticum pilus | | Descriptor: | pilin | | Authors: | Wang, F, Baquero, D.P, Su, Z, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The structures of two archaeal type IV pili illuminate evolutionary relationships.
Nat Commun, 11, 2020
|
|
6W8X
 
 | | Cryo-EM of the S. solfataricus pilus | | Descriptor: | pilin | | Authors: | Wang, F, Baquero, D.P, Su, Z, Beltran, L.C, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2020-03-21 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structures of two archaeal type IV pili illuminate evolutionary relationships.
Nat Commun, 11, 2020
|
|
6VY1
 
 | | Cryo-EM structure of filamentous PFD from Methanocaldococcus jannaschii | | Descriptor: | Prefoldin subunit alpha 2 | | Authors: | Wang, F, Chen, Y.X, Ing, N.L, Hochbaum, A.I, Clark, D.S, Glover, D.J, Egelman, E.H. | | Deposit date: | 2020-02-25 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structural Determination of a Filamentous Chaperone to Fabricate Electronically Conductive Metalloprotein Nanowires.
Acs Nano, 14, 2020
|
|
6WL1
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3 | | Descriptor: | peptide 36-31-3 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL8
 
 | | Cryo-EM of Form 2 peptide filament | | Descriptor: | Form 2 peptide | | Authors: | Wang, F, Gnewou, O.M, Xu, C, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WL0
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3-RD | | Descriptor: | peptide 36-31-3-RD | | Authors: | Wang, F, Gnewou, O.M, Su, Z, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
4RL1
 
 | |
6V7B
 
 | | Cryo-EM reconstruction of Pyrobaculum filamentous virus 2 (PFV2) | | Descriptor: | A-DNA, Structural protein VP1, Structural protein VP2 | | Authors: | Wang, F, Baquero, D.P, Su, Z, Prangishvili, D, Krupovic, M, Egelman, E.H. | | Deposit date: | 2019-12-08 | | Release date: | 2020-04-01 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of a filamentous virus uncovers familial ties within the archaeal virosphere.
Virus Evol, 6, 2020
|
|
8FJ5
 
 | | Structure of the Haloferax volcanii archaeal type IV pilus | | Descriptor: | Pilin_N domain-containing protein | | Authors: | Wang, F, Kreutzberger, M.A, Baquero, D.P, Krupovic, M, Egelman, E.H. | | Deposit date: | 2022-12-19 | | Release date: | 2023-06-28 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | The evolution of archaeal flagellar filaments.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5EJL
 
 | | MrkH, A novel c-di-GMP dependence transcription regulatory factor. | | Descriptor: | 1,2-ETHANEDIOL, 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Klebsiella pneumoniae genome assembly NOVST, ... | | Authors: | Wang, F, Zhu, D, Gu, L. | | Deposit date: | 2015-11-02 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The PilZ domain of MrkH represents a novel DNA binding motif
Protein Cell, 7, 2016
|
|
8WTF
 
 | | The Crystal Structure of IRAK4 from Biortus | | Descriptor: | 1,2-ETHANEDIOL, 1-{[(2S,3S,4S)-3-ethyl-4-fluoro-5-oxopyrrolidin-2-yl]methoxy}-7-methoxyisoquinoline-6-carboxamide, Interleukin-1 receptor-associated kinase 4, ... | | Authors: | Wang, F, Cheng, W, Lv, Z, Meng, Q, Xu, Y. | | Deposit date: | 2023-10-18 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of IRAK4 from Biortus
To Be Published
|
|
8WFQ
 
 | | The Crystal Structure of RALDH1 from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Aldehyde dehydrogenase 1A1 | | Authors: | Wang, F, Cheng, W, Lv, Z, Qi, J, Shen, Z. | | Deposit date: | 2023-09-20 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The Crystal Structure of RALDH1 from Biortus.
To Be Published
|
|
5GWZ
 
 | | The structure of Porcine epidemic diarrhea virus main protease in complex with an inhibitor | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, PEDV main protease | | Authors: | Wang, F, Chen, C, Yang, K, Liu, X, Liu, H, Xu, Y, Chen, X, Liu, X, Cai, Y, Yang, H. | | Deposit date: | 2016-09-14 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.444 Å) | | Cite: | Michael Acceptor-Based Peptidomimetic Inhibitor of Main Protease from Porcine Epidemic Diarrhea Virus
J. Med. Chem., 60, 2017
|
|
2HKC
 
 | | NMR Structure of the IQ-modified Dodecamer CTCGGC[IQ]GCCATC | | Descriptor: | 3-METHYL-3H-IMIDAZO[4,5-F]QUINOLIN-2-AMINE, 5'-D(*CP*TP*CP*GP*GP*CP*GP*CP*CP*AP*TP*C)-3', 5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3' | | Authors: | Wang, F, DeMuro, N.E, Elmquist, C.E, Stover, J.S, Rizzo, C.J, Stone, M.P. | | Deposit date: | 2006-07-03 | | Release date: | 2006-10-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Base-displaced intercalated structure of the food mutagen 2-amino-3-methylimidazo[4,5-f]quinoline in the recognition sequence of the NarI restriction enzyme, a hotspot for -2 bp deletions.
J.Am.Chem.Soc., 128, 2006
|
|
5EEG
 
 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with tetrazole-SAH | | Descriptor: | (2~{R},3~{R},4~{S},5~{S})-2-(6-aminopurin-9-yl)-5-[[(3~{S})-3-azanyl-3-(1~{H}-1,2,3,4-tetrazol-5-yl)propyl]sulfanylmethyl]oxolane-3,4-diol, Carminomycin 4-O-methyltransferase DnrK | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.255 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
5D6X
 
 | | Crystal structure of double tudor domain of human lysine demethylase KDM4A | | Descriptor: | Lysine-specific demethylase 4A, SULFATE ION | | Authors: | Wang, F, Su, Z, Denu, J.M, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-08-13 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.153 Å) | | Cite: | Reader domain specificity and lysine demethylase-4 family function.
Nat Commun, 7, 2016
|
|
5EEH
 
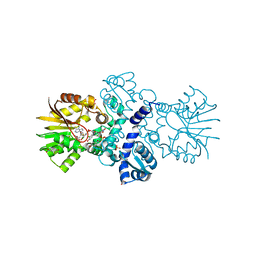 | | Crystal structure of carminomycin-4-O-methyltransferase DnrK in complex with SAH and 2-chloro-4-nitrophenol | | Descriptor: | 2-chloranyl-4-nitro-phenol, Carminomycin 4-O-methyltransferase DnrK, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Wang, F, Singh, S, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-10-22 | | Release date: | 2015-12-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Functional AdoMet Isosteres Resistant to Classical AdoMet Degradation Pathways.
Acs Chem.Biol., 11, 2016
|
|
8UMT
 
 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the three-subunit binding state (state 2) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UNJ
 
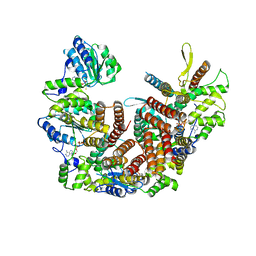 | | Atomic model of the human CTF18-RFC alone in the apo state (State 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8UMU
 
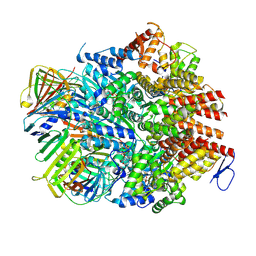 | | Atomic model of the human CTF18-RFC-PCNA binary complex in the four-subunit binding state (state 3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chromosome transmission fidelity protein 18 homolog, MAGNESIUM ION, ... | | Authors: | Wang, F, He, Q, Li, H. | | Deposit date: | 2023-10-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM reveals a nearly complete PCNA loading process and unique features of the human alternative clamp loader CTF18-RFC.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
