8JED
 
 | | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors | | Descriptor: | mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Wang, D, Zhao, R, Shu, W, Hu, W, Wang, M, Cao, J, Zhou, X. | | Deposit date: | 2023-05-15 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure of mRNA cap (guanine-N7) methyltransferase E12 subunit from monkeypox virus and discovery of its inhibitors.
Int.J.Biol.Macromol., 253, 2023
|
|
8YYR
 
 | | Structure of the HitB T293G mutant | | Descriptor: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(2-bromophenyl)propanoyl]sulfamate | | Authors: | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2024-04-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
8YYQ
 
 | | Structure of the HitB F328L mutant | | Descriptor: | Putative ATP-dependent b-aminoacyl-ACP synthetase, [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl ~{N}-[(3~{S})-3-azanyl-3-(3-cyanophenyl)propanoyl]sulfamate | | Authors: | Wang, D, Miyanaga, A, Chisuga, T, Kudo, F, Eguchi, T. | | Deposit date: | 2024-04-04 | | Release date: | 2024-06-05 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Engineering the Substrate Specificity of (S)-beta-Phenylalanine Adenylation Enzyme HitB.
Chembiochem, 25, 2024
|
|
7XT0
 
 | | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors | | Descriptor: | 1,2-ETHANEDIOL, RNA helicase | | Authors: | Wang, D.P, Jiang, F.Y, Zeng, X.Y, Zhao, R, Chen, C, Zhu, Y, Cao, J.M. | | Deposit date: | 2022-05-15 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Crystal structure of RNA helicase from Saint Louis encephalitis virus and discovery of its inhibitors
To Be Published
|
|
1SNB
 
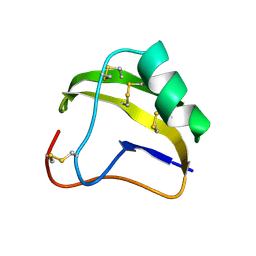 | | STRUCTURE OF SCORPION NEUROTOXIN BMK M8 | | Descriptor: | NEUROTOXIN BMK M8 | | Authors: | Wang, D.C, Zeng, Z.H, Li, H.M. | | Deposit date: | 1997-03-12 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an acidic neurotoxin from scorpion Buthus martensii Karsch at 1.85 A resolution.
J.Mol.Biol., 261, 1996
|
|
4WIL
 
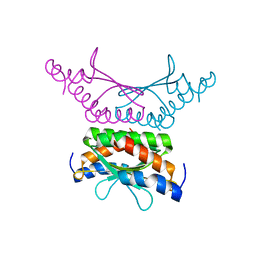 | | Crystal structure of DCoH2 S51T | | Descriptor: | Pterin-4-alpha-carbinolamine dehydratase 2 | | Authors: | Wang, D, Rose, R.B. | | Deposit date: | 2014-09-26 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Interactions with the Bifunctional Interface of the Transcriptional Coactivator DCoH1 Are Kinetically Regulated.
J.Biol.Chem., 290, 2015
|
|
1S9K
 
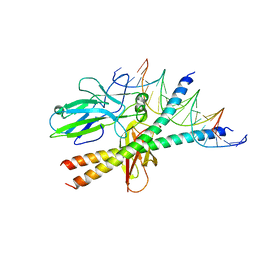 | |
7WD4
 
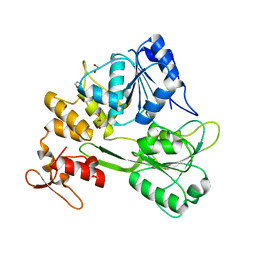 | | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, NS3 helicase | | Authors: | Wang, D.P, Wang, M.Y, Zhou, X, Wang, W.M, Cao, J.M. | | Deposit date: | 2021-12-21 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the Ilheus virus helicase: implications for enzyme function and drug design
To Be Published
|
|
3OSV
 
 | | The crytsal structure of FLGD from P. Aeruginosa | | Descriptor: | Flagellar basal-body rod modification protein FlgD, GLYCEROL | | Authors: | Wang, D, Luo, M, Niu, S. | | Deposit date: | 2010-09-10 | | Release date: | 2011-05-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of a novel dimer form of FlgD from P. aeruginosa PAO1
Proteins, 79, 2011
|
|
2CGA
 
 | |
4FF5
 
 | |
2E2J
 
 | | RNA polymerase II elongation complex in 5 mM Mg+2 with GMPCPP | | Descriptor: | 27-MER DNA template strand, 5'-D(P*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*A)-3', 5'-R(P*AP*UP*CP*GP*AP*GP*AP*GP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2E2H
 
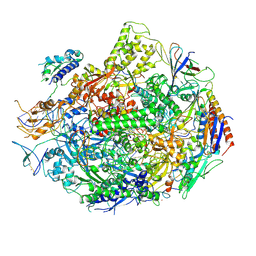 | | RNA polymerase II elongation complex at 5 mM Mg2+ with GTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
5CRL
 
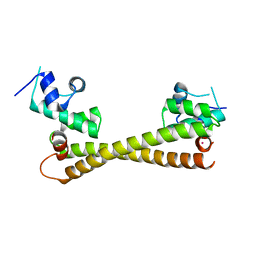 | |
2E2I
 
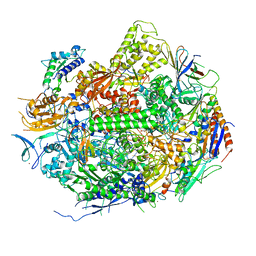 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dGTP | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
3GTM
 
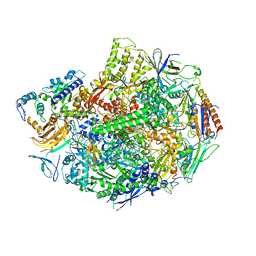 | | Co-complex of Backtracked RNA polymerase II with TFIIS | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3M3Y
 
 | | RNA polymerase II elongation complex C | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M4O
 
 | | RNA polymerase II elongation complex B | | Descriptor: | DNA (28-MER), DNA (5'-D(P*GP*TP*GP*GP*TP*TP*AP*TP*GP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Zhu, G, Huang, X, Lippard, S.J. | | Deposit date: | 2010-03-11 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | X-ray structure and mechanism of RNA polymerase II stalled at an antineoplastic monofunctional platinum-DNA adduct.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5X8Y
 
 | | A Mutation identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in vitro | | Descriptor: | ZIKV NS1 | | Authors: | Wang, D, Chen, C, Liu, S, Zhou, H, Yang, K, Zhao, Q, Ji, X, Chen, C, Xie, W, Wang, Z, Mi, L.Z, Yang, H. | | Deposit date: | 2017-03-03 | | Release date: | 2017-05-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.817 Å) | | Cite: | A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro
Sci Rep, 7, 2017
|
|
3GTL
 
 | | Backtracked RNA polymerase II complex with 13mer with G<>U mismatch | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTK
 
 | | Backtracked RNA polymerase II complex with 18mer RNA | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTP
 
 | | Backtracked RNA polymerase II complex with 24mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
5GSZ
 
 | | Crystal Structure of the KIF19A Motor Domain Complexed with Mg-ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF19, MAGNESIUM ION | | Authors: | Wang, D, Nitta, R, Hirokawa, N. | | Deposit date: | 2016-08-18 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A
Elife, 5, 2016
|
|
3GTG
 
 | | Backtracked RNA polymerase II complex with 12mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.78 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
3GTJ
 
 | | Backtracked RNA polymerase II complex with 13mer RNA | | Descriptor: | DNA (28-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, D, Bushnell, D.A, Huang, X, Westover, K.D, Levitt, M, Kornberg, R.D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural basis of transcription: backtracked RNA polymerase II at 3.4 angstrom resolution.
Science, 324, 2009
|
|
