7SQG
 
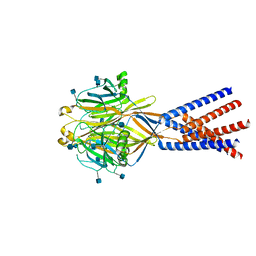 | |
6VYQ
 
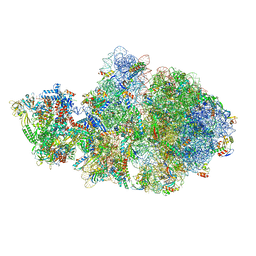 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing an 15 nt long mRNA spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZ7
 
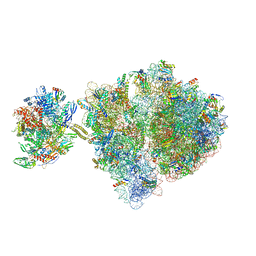 | | Escherichia coli transcription-translation complex C1 (TTC-C1) containing a 27 nt long mRNA spacer, NusG, and fMet-tRNAs at P-site and E-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
2VGF
 
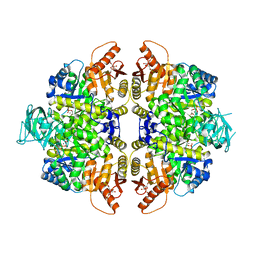 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: T384M mutant | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L.R, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-12 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
2V7Z
 
 | | Crystal structure of the 70-kDa heat shock cognate protein from Rattus norvegicus in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT SHOCK COGNATE 71 KDA PROTEIN, PHOSPHATE ION | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
6W8T
 
 | | Crystal structure of metacaspase 4 from Arabidopsis (microcrystals treated with calcium) | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
6VU3
 
 | | Cryo-EM structure of Escherichia coli transcription-translation complex A (TTC-A) containing mRNA with a 12 nt long spacer | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R. | | Deposit date: | 2020-02-14 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VZJ
 
 | | Escherichia coli transcription-translation complex A1 (TTC-A1) containing mRNA with a 15 nt long spacer, fMet-tRNAs at E-site and P-site, and lacking transcription factor NusG | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-28 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6W8S
 
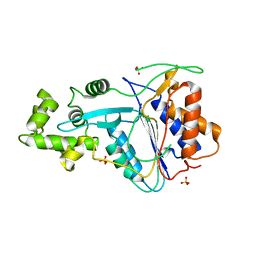 | | Crystal structure of metacaspase 4 from Arabidopsis | | Descriptor: | Metacaspase-4, SULFATE ION | | Authors: | Zhu, P, Yu, X.H, Wang, C, Zhang, Q, Liu, W, McSweeney, S, Shanklin, J, Lam, E, Liu, Q. | | Deposit date: | 2020-03-21 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.484 Å) | | Cite: | Structural basis for Ca2+-dependent activation of a plant metacaspase.
Nat Commun, 11, 2020
|
|
2V7Y
 
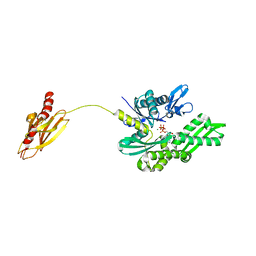 | | Crystal structure of the molecular chaperone DnaK from Geobacillus kaustophilus HTA426 in post-ATP hydrolysis state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPERONE PROTEIN DNAK, MAGNESIUM ION, ... | | Authors: | Chang, Y.-W, Sun, Y.-J, Wang, C, Hsiao, C.-D. | | Deposit date: | 2007-08-02 | | Release date: | 2008-04-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal Structures of the 70-kDa Heat Shock Proteins in Domain Disjoining Conformation.
J.Biol.Chem., 283, 2008
|
|
6VZ5
 
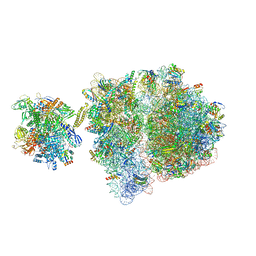 | | Escherichia coli transcription-translation complex D3 (TTC-D3) containing mRNA with a 21 nt long spacer, NusG, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6VYZ
 
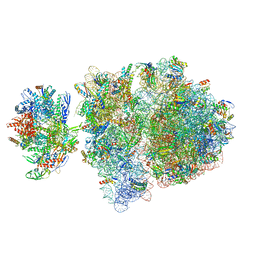 | | Escherichia coli transcription-translation complex C6 (TTC-C6) containing mRNA with a 21 nt long spacer, NusA, and fMet-tRNAs at E-site and P-site | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S1, ... | | Authors: | Molodtsov, V, Wang, C, Su, M, Ebright, R.H. | | Deposit date: | 2020-02-27 | | Release date: | 2020-09-02 | | Last modified: | 2020-09-23 | | Method: | ELECTRON MICROSCOPY (9.9 Å) | | Cite: | Structural basis of transcription-translation coupling.
Science, 369, 2020
|
|
6NZK
 
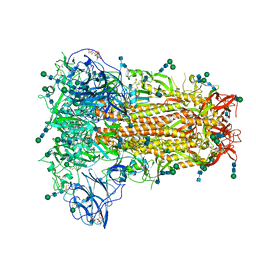 | | Structural basis for human coronavirus attachment to sialic acid receptors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-02-13 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
2VGI
 
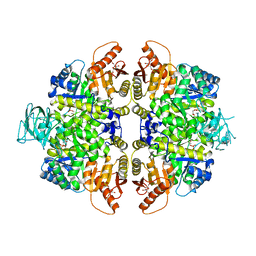 | | HUMAN ERYTHROCYTE PYRUVATE KINASE: R486W MUTANT | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 2-PHOSPHOGLYCOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Valentini, G, Chiarelli, L, Fortin, R, Dolzan, M, Galizzi, A, Abraham, D.J, Wang, C, Bianchi, P, Zanella, A, Mattevi, A. | | Deposit date: | 2007-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Structure and Function of Human Erythrocyte Pyruvate Kinase. Molecular Basis of Nonspherocytic Hemolytic Anemia.
J.Biol.Chem., 277, 2002
|
|
6OHW
 
 | | Structural basis for human coronavirus attachment to sialic acid receptors. Apo-HCoV-OC43 S | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike surface glycoprotein, ... | | Authors: | Tortorici, M.A, Walls, A.C, Lang, Y, Wang, C, Li, Z, Koerhuis, D, Boons, G.J, Bosch, B.J, Rey, F.A, de Groot, R, Veesler, D. | | Deposit date: | 2019-04-07 | | Release date: | 2019-06-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human coronavirus attachment to sialic acid receptors.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6DNF
 
 | |
2WPV
 
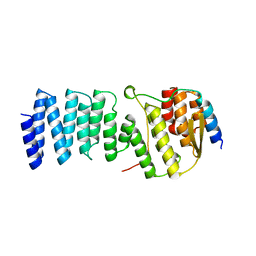 | | Crystal structure of S. cerevisiae Get4-Get5 complex | | Descriptor: | MERCURY (II) ION, UBIQUITIN-LIKE PROTEIN MDY2, UPF0363 PROTEIN YOR164C | | Authors: | Chang, Y.-W, Chuang, Y.-C, Ho, Y.-C, Cheng, M.-Y, Sun, Y.-J, Hsiao, C.-D, Wang, C. | | Deposit date: | 2009-08-11 | | Release date: | 2010-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of Get4-Get5 Complex and its Interactions with Sgt2, Get3, and Ydj1.
J.Biol.Chem., 285, 2010
|
|
3WUY
 
 | | Crystal structure of Nit6803 | | Descriptor: | Nitrilase | | Authors: | Yuan, Y.A, Yin, B, Wang, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural insights into enzymatic activity and substrate specificity determination by a single amino acid in nitrilase from Syechocystis sp. PCC6803
J.Struct.Biol., 188, 2014
|
|
1AUV
 
 | |
1AUX
 
 | |
8WJY
 
 | | PKMYT1_Cocrystal_Cpd 4 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Membrane-associated tyrosine- and threonine-specific cdc2-inhibitory kinase, ... | | Authors: | Wang, Y, Wang, C, Liu, T, Qi, H, Chen, S, Cai, X, Zhang, M, Aliper, A, Ren, F, Ding, X, Zhavoronkov, A. | | Deposit date: | 2023-09-26 | | Release date: | 2023-11-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of Tetrahydropyrazolopyrazine Derivatives as Potent and Selective MYT1 Inhibitors for the Treatment of Cancer.
J.Med.Chem., 67, 2024
|
|
8IYD
 
 | | Tail cap of phage lambda tail | | Descriptor: | Tail tube protein, Tail tube terminator protein | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-04 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYK
 
 | | Tail tip conformation 1 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
8IYL
 
 | | Tail tip conformation 2 of phage lambda tail | | Descriptor: | IRON/SULFUR CLUSTER, Tail tip assembly protein I, Tail tip protein L, ... | | Authors: | Wang, J.W, Wang, C. | | Deposit date: | 2023-04-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the bacteriophage lambda tail.
Structure, 32, 2024
|
|
3LD8
 
 | | Structure of JMJD6 and Fab Fragments | | Descriptor: | Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, FE (III) ION, GLYCEROL, ... | | Authors: | Hong, X, Zang, J, White, J, Kappler, J.W, Wang, C, Zhang, G. | | Deposit date: | 2010-01-12 | | Release date: | 2010-08-04 | | Last modified: | 2012-06-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Interaction of JMJD6 with single-stranded RNA.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
