1O06
 
 | | Crystal structure of the Vps27p Ubiquitin Interacting Motif (UIM) | | Descriptor: | Vacuolar protein sorting-associated protein VPS27, ZINC ION | | Authors: | Fisher, R.D, Wang, B, Alam, S.L, Higginson, D.S, Rich, R, Myszka, D, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2003-02-20 | | Release date: | 2003-07-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and ubiquitin binding of the ubiquitin-interacting motif.
J.Biol.Chem., 278, 2003
|
|
3HEI
 
 | | Ligand Recognition by A-Class Eph Receptors: Crystal Structures of the EphA2 Ligand-Binding Domain and the EphA2/ephrin-A1 Complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1 | | Authors: | Himanen, J.P, Goldgur, Y, Miao, H, Myshkin, E, Guo, H, Buck, M, Nguyen, M, Rajashankar, K.R, Wang, B, Nikolov, D.B. | | Deposit date: | 2009-05-08 | | Release date: | 2009-06-30 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Ligand recognition by A-class Eph receptors: crystal structures of the EphA2 ligand-binding domain and the EphA2/ephrin-A1 complex.
Embo Rep., 10, 2009
|
|
2P5Y
 
 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
2P8T
 
 | | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0730 | | Authors: | Chen, L, Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3
To be Published
|
|
2P17
 
 | | Crystal structure of GK1651 from Geobacillus kaustophilus | | Descriptor: | FE (III) ION, Pirin-like protein | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of GK1651 from Geobacillus kaustophilus
To be Published
|
|
2PHC
 
 | | Crystal structure of conserved uncharacterized protein PH0987 from Pyrococcus horikoshii | | Descriptor: | Uncharacterized protein PH0987 | | Authors: | Swindell II, J.T, Chen, L, Zhu, J, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-10 | | Release date: | 2007-05-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of conserved uncharacterized protein PH0987 from Pyrococcus horikoshii.
To be Published
|
|
2PW6
 
 | | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12 | | Descriptor: | Uncharacterized protein ygiD, ZINC ION | | Authors: | Newton, M.G, Takagi, Y, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokayama, S, Li, Y, Chen, L, Zhu, J, Ruble, J, Liu, Z.J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-10 | | Release date: | 2007-06-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal structure of uncharacterized protein JW3007 from Escherichia coli K12.
To be Published
|
|
2QVO
 
 | | Crystal structure of AF1382 from Archaeoglobus fulgidus | | Descriptor: | Uncharacterized protein AF_1382 | | Authors: | Zhu, J, Zhao, M, Fu, Z.-Q, Yang, H, Chang, J, Hao, X, Chen, L, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-08-08 | | Release date: | 2007-09-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of AF1382 from Archaeoglobus fulgidus.
To be Published
|
|
5U8W
 
 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa bound to NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, ... | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
2DST
 
 | | Crystal Structure Analysis of TT1977 | | Descriptor: | Hypothetical protein TTHA1544 | | Authors: | Xie, Y, Kishishita, S, Murayama, K, Shirouzu, M, Chen, L, Liu, Z.J, Wang, B.C, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-07-06 | | Release date: | 2007-01-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the minimized alpha/beta-hydrolase fold protein from Thermus thermophilus HB8.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
4PGO
 
 | | Crystal structure of hypothetical protein PF0907 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PII
 
 | | Crystal structure of hypothetical protein PF0907 from pyrococcus furiosus solved by sulfur SAD using Swiss light source data | | Descriptor: | CHLORIDE ION, IMIDAZOLE, N-glycosylase/DNA lyase | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-05-08 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2EJ5
 
 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
5U8U
 
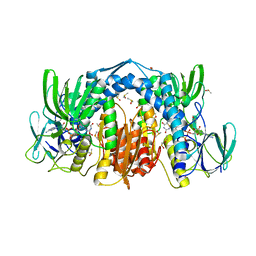 | | Dihydrolipoamide dehydrogenase (LpdG) from Pseudomonas aeruginosa | | Descriptor: | DIMETHYL SULFOXIDE, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Glasser, N.R, Wang, B.X, Hoy, J.A, Newman, D.K. | | Deposit date: | 2016-12-15 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Pyruvate and alpha-Ketoglutarate Dehydrogenase Complexes of Pseudomonas aeruginosa Catalyze Pyocyanin and Phenazine-1-carboxylic Acid Reduction via the Subunit Dihydrolipoamide Dehydrogenase.
J. Biol. Chem., 292, 2017
|
|
2DYY
 
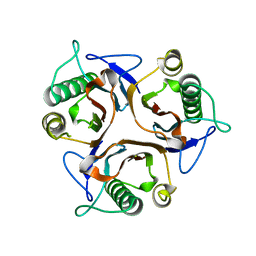 | | Crystal structure of putative translation initiation inhibitor PH0854 from Pyrococcus horikoshii | | Descriptor: | UPF0076 protein PH0854 | | Authors: | Ihsanawati, Kishishita, S, Murayama, K, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Bessho, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-19 | | Release date: | 2007-03-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of putative translation initiation inhibitor PH0854 from Pyrococcus horikoshii
To be Published
|
|
3GED
 
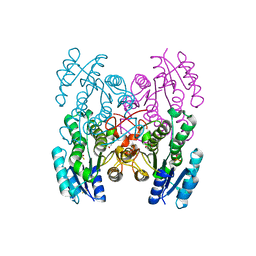 | | Fingerprint and Structural Analysis of a Apo SCOR enzyme from Clostridium thermocellum | | Descriptor: | GLYCEROL, SODIUM ION, Short-chain dehydrogenase/reductase SDR, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
3GEG
 
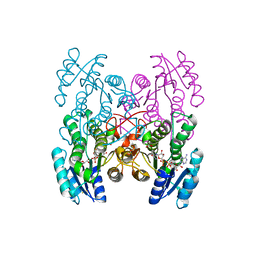 | | Fingerprint and Structural Analysis of a SCOR enzyme with its bound cofactor from Clostridium thermocellum | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Huether, R, Liu, Z.J, Xu, H, Wang, B.C, Pletnev, V, Mao, Q, Umland, T, Duax, W. | | Deposit date: | 2009-02-25 | | Release date: | 2009-03-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Sequence fingerprint and structural analysis of the SCOR enzyme A3DFK9 from Clostridium thermocellum.
Proteins, 78, 2010
|
|
7JSJ
 
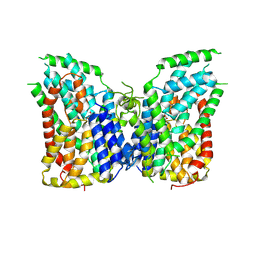 | | Structure of the NaCT-PF2 complex | | Descriptor: | (2R)-2-[2-(4-tert-butylphenyl)ethyl]-2-hydroxybutanedioic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
7JSK
 
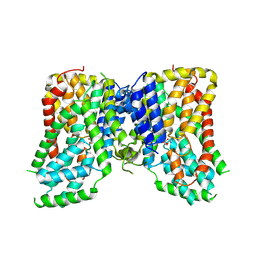 | | Structure of the NaCT-Citrate complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, SODIUM ION, ... | | Authors: | Sauer, D.B, Wang, B, Song, J, Rice, W.J, Wang, D.N. | | Deposit date: | 2020-08-14 | | Release date: | 2021-02-24 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structure and inhibition mechanism of the human citrate transporter NaCT.
Nature, 591, 2021
|
|
4TN8
 
 | | Crystal structure of Thermus Thermophilus thioredoxin solved by sulfur SAD using Swiss Light Source data | | Descriptor: | CHLORIDE ION, Thioredoxin | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-03 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4TNO
 
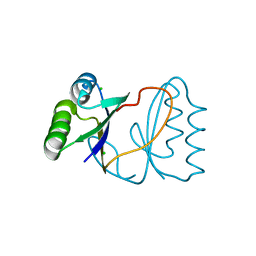 | | Hypothetical protein PF1117 from Pyrococcus Furiosus: Structure solved by sulfur-SAD using Swiss Light Source Data | | Descriptor: | CHLORIDE ION, CRISPR-associated endoribonuclease Cas2 | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-06-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
2HIQ
 
 | | Crystal structure of JW1657 from Escherichia coli | | Descriptor: | Hypothetical protein ydhR | | Authors: | Chen, L.Q, Chen, L.R, Liu, Z.-J, Temple, W, Lee, D, Chang, S.-H, Rose, J.P, Ebihara, A, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-09-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of JW1657 from Escherichia coli at 2.0A resolution
To be Published
|
|
2HQ4
 
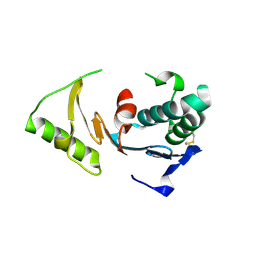 | | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii | | Descriptor: | Hypothetical protein PH1570 | | Authors: | Li, Y, Marshall, M, Chang, J, Zhao, M, Zhang, M, Xu, H, Liu, Z.J, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2006-07-18 | | Release date: | 2006-09-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of ORF 1580 a hypothetical protein from Pyrococcus horikoshii
To be Published
|
|
1UT9
 
 | | Structural Basis for the Exocellulase Activity of the Cellobiohydrolase CbhA from C. thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Schubot, F.D, Kataeva, I.A, Chang, J, Shah, A.K, Ljungdahl, L.G, Rose, J.P, Wang, B.C. | | Deposit date: | 2003-12-04 | | Release date: | 2004-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the exocellulase activity of the cellobiohydrolase CbhA from Clostridium thermocellum.
Biochemistry, 43, 2004
|
|
1VJK
 
 | | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 | | Descriptor: | molybdopterin converting factor, subunit 1 | | Authors: | Chen, L, Liu, Z.J, Tempel, W, Shah, A, Lee, D, Rose, J.P, Eneh, J.C, Hopkins, R.C, Jenney Jr, F.E, Lee, H.S, Li, T, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-03-10 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Putative molybdopterin converting factor, subunit 1 from Pyrococcus furiosus, Pfu-562899-001 '
To be published
|
|
