5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
6QGK
 
 | | Structure of human Bcl-2 in complex with THIQ-phenyl pyrazole compound | | Descriptor: | 1-[2-[[(3~{S})-3-(aminomethyl)-3,4-dihydro-1~{H}-isoquinolin-2-yl]carbonyl]phenyl]-~{N},~{N}-dibutyl-5-methyl-pyrazole-3-carboxamide, ACETATE ION, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1 | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
3V71
 
 | | Crystal structure of PUF-6 in complex with 5BE13 RNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Puf (Pumilio/fbf) domain-containing protein 7, confirmed by transcript evidence, ... | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
6QGD
 
 | | Structure of human Mcl-1 in complex with thienopyrimidine inhibitor | | Descriptor: | 2-[(6-ethyl-5-phenyl-thieno[2,3-d]pyrimidin-4-yl)amino]-3-oxidanyl-propanoic acid, Maltose-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, SODIUM ION, ... | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6QGG
 
 | | Structure of human Bcl-2 in complex with analogue of ABT-737 | | Descriptor: | Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2,Bcl-2-like protein 1, [(3~{R})-3-[[4-[[4-[4-[[2-(4-chlorophenyl)phenyl]methyl]piperazin-1-yl]phenyl]carbonylsulfamoyl]-2-nitro-phenyl]amino]-4-phenylsulfanyl-butyl]-(2-hydroxy-2-oxoethyl)-dimethyl-azanium | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-11 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
3V6Y
 
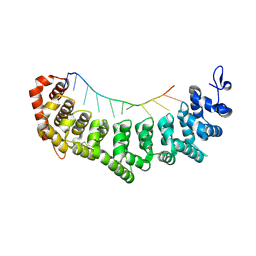 | | crystal structure of FBF-2 in complex with a mutant gld-1 FBEa13 RNA | | Descriptor: | Fem-3 mRNA-binding factor 2, RNA (5'-R(*UP*AP*CP*UP*GP*UP*GP*CP*CP*AP*UP*AP*C)-3') | | Authors: | Qiu, C, Kershner, A, Wang, Y, Holley, C.H, Wilinski, D, Keles, S, Kimble, J, Wickens, M, Hall, T.M.T. | | Deposit date: | 2011-12-20 | | Release date: | 2012-01-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Divergence of PUF protein specificity through variations in an RNA-binding pocket
J.Biol.Chem., 2012
|
|
5KLN
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169A in complex with NAD+ | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLK
 
 | | Crystal structure of 2-aminomuconate 6-semialdehyde dehydrogenase N169D in complex with NAD+ and 2-hydroxymuconate-6-semialdehyde | | Descriptor: | (2E,4E)-2-hydroxy-6-oxohexa-2,4-dienoic acid, 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
4W9P
 
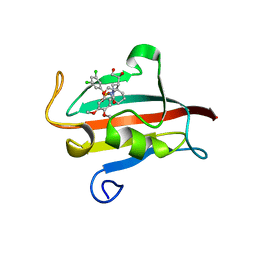 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-5-[(1S)-1,2-dihydroxyethyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-3,10-diazabicyclo[4.3.1]decan-2-one, ACETATE ION, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1RXT
 
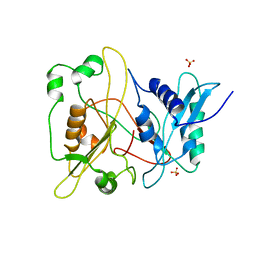 | | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase. | | Descriptor: | COBALT (II) ION, Glycylpeptide N-tetradecanoyltransferase 1, SULFATE ION | | Authors: | Yang, J, Wang, Y, Frey, G, Abeles, R.H, Petsko, G.A, Ringe, D. | | Deposit date: | 2003-12-18 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human myristoyl-CoA:protein N-myristoyltransferase
To be Published
|
|
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1S8K
 
 | | Solution Structure of BmKK4, A Novel Potassium Channel Blocker from Scorpion Buthus martensii Karsch, 25 structures | | Descriptor: | Toxin BmKK4 | | Authors: | Zhang, N, Chen, X, Li, M, Cao, C, Wang, Y, Hu, G, Wu, H. | | Deposit date: | 2004-02-02 | | Release date: | 2005-02-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of BmKK4, the first member of subfamily alpha-KTx 17 of scorpion toxins
Biochemistry, 43, 2004
|
|
5W1L
 
 | |
6J75
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase | | Descriptor: | Aldehyde dehydrogenase A | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.695 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
3CIG
 
 | | Crystal structure of mouse TLR3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-11 | | Release date: | 2008-05-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
7BXG
 
 | | MavC-UBE2N-Ub complex | | Descriptor: | MavC, Polyubiquitin-C, Ubiquitin-conjugating enzyme E2 N | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.705 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
7BXH
 
 | | MavC-Lpg2149 complex | | Descriptor: | Lpg2149, MavC | | Authors: | Gao, P, Wang, Y. | | Deposit date: | 2020-04-19 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into catalysis and regulation of non-canonical ubiquitination and deubiquitination by bacterial deamidase effectors.
Nat Commun, 11, 2020
|
|
3CPU
 
 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
6K3B
 
 | | Crystal structure of Lpg2147-Lpg2149 complex | | Descriptor: | Lpg2147, Uncharacterized protein | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-05-17 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.974 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
3D7A
 
 | |
3CIY
 
 | | Mouse Toll-like receptor 3 ectodomain complexed with double-stranded RNA | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, L, Botos, I, Wang, Y, Leonard, J.N, Shiloach, J, Segal, D.M, Davies, D.R. | | Deposit date: | 2008-03-12 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Structural basis of toll-like receptor 3 signaling with double-stranded RNA.
Science, 320, 2008
|
|
1D3K
 
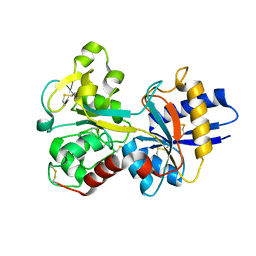 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, SERUM TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-09-29 | | Release date: | 2000-03-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
6KFP
 
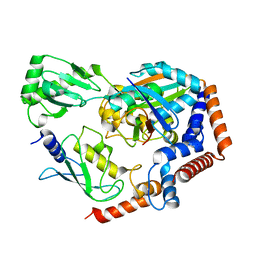 | | Crystal structure of MavC ternary complex | | Descriptor: | MavC, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 N | | Authors: | Mu, Y, Wang, Y, Han, Y, Li, D, Feng, Y. | | Deposit date: | 2019-07-08 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural insights into the mechanism and inhibition of transglutaminase-induced ubiquitination by the Legionella effector MavC.
Nat Commun, 11, 2020
|
|
1D4N
 
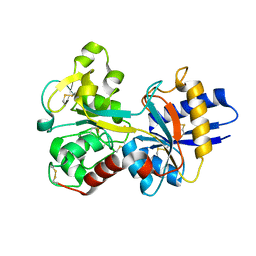 | | HUMAN SERUM TRANSFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, TRANSFERRIN | | Authors: | Yang, H.-W, MacGillivray, R.T.A, Chen, J, Luo, Y, Wang, Y, Brayer, G.D, Mason, A, Woodworth, R.C, Murphy, M.E.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of two mutants (K206Q, H207E) of the N-lobe of human transferrin with increased affinity for iron.
Protein Sci., 9, 2000
|
|
6JFU
 
 | | Crystal structure of Nme2Cas9 in complex with sgRNA and target DNA (AGGCCC PAM) | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9, non-target strand, ... | | Authors: | Sun, W, Yang, J, Cheng, Z, Liu, C, Wang, K, Huang, X, Wang, Y. | | Deposit date: | 2019-02-12 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Neisseria meningitidis Cas9 Complexes in Catalytically Poised and Anti-CRISPR-Inhibited States.
Mol.Cell, 76, 2019
|
|
