7D5I
 
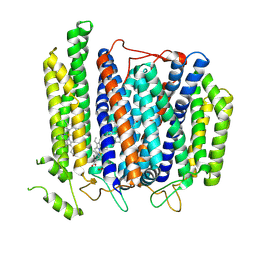 | | Structure of Mycobacterium smegmatis bd complex in the apo-form. | | 分子名称: | CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome D ubiquinol oxidase subunit 1, HEME B/C, ... | | 著者 | Wang, W, Gong, H, Gao, Y, Zhou, X, Rao, Z. | | 登録日 | 2020-09-26 | | 公開日 | 2021-06-23 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (2.79 Å) | | 主引用文献 | Cryo-EM structure of mycobacterial cytochrome bd reveals two oxygen access channels.
Nat Commun, 12, 2021
|
|
7VP8
 
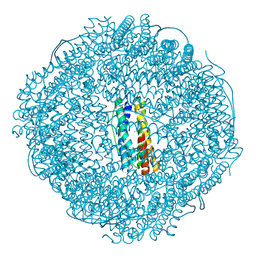 | | Crystal structure of ferritin from Ureaplasma urealyticum | | 分子名称: | CHLORIDE ION, FE (III) ION, Ferritin-like diiron domain-containing protein | | 著者 | Wang, W, Liu, X, Wang, Y, Fu, D, Wang, H. | | 登録日 | 2021-10-15 | | 公開日 | 2022-08-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | Distinct structural characteristics define a new subfamily of Mycoplasma ferritin
Chin.Chem.Lett., 33, 2022
|
|
7VF4
 
 | | Crystal structure of Vps75 from Candida albicans | | 分子名称: | CHLORIDE ION, SODIUM ION, Vps75 | | 著者 | Wang, W, Chen, X, Yang, Z, Chen, X, Li, C, Wang, M. | | 登録日 | 2021-09-10 | | 公開日 | 2021-10-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Crystal structure of histone chaperone Vps75 from Candida albicans.
Biochem.Biophys.Res.Commun., 578, 2021
|
|
7TO1
 
 | |
7TO0
 
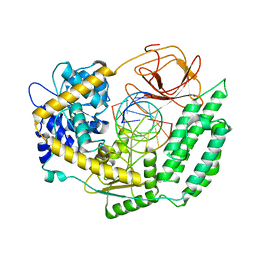 | | Cryo-EM structure of RIG-I in complex with OHdsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, OHdsRNA, ZINC ION | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.5 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNZ
 
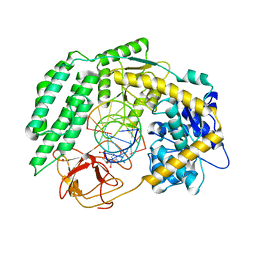 | | Cryo-EM structure of RIG-I in complex with p1dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p1dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TNY
 
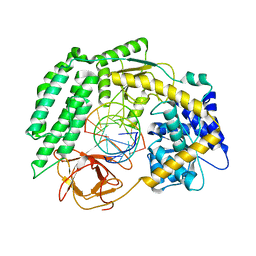 | | Cryo-EM structure of RIG-I in complex with p2dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p2dsRNA | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
7TO2
 
 | |
7TNX
 
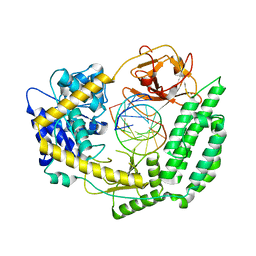 | | Cryo-EM structure of RIG-I in complex with p3dsRNA | | 分子名称: | Antiviral innate immune response receptor RIG-I, ZINC ION, p3dsRNAa, ... | | 著者 | Wang, W, Pyle, A.M. | | 登録日 | 2022-01-22 | | 公開日 | 2022-11-02 | | 最終更新日 | 2024-06-05 | | 実験手法 | ELECTRON MICROSCOPY (3.54 Å) | | 主引用文献 | The RIG-I receptor adopts two different conformations for distinguishing host from viral RNA ligands.
Mol.Cell, 82, 2022
|
|
6Z5J
 
 | | Arrangement of the matrix protein M1 in influenza A/Hong Kong/1/1968 VLPs (HA,NA,M1,M2) | | 分子名称: | Matrix protein 1 | | 著者 | Peukes, J, Xiong, X, Erlendsson, S, Qu, K, Wan, W, Kraeusslich, H.-G, Briggs, J.A.G. | | 登録日 | 2020-05-26 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (8 Å) | | 主引用文献 | The native structure of the assembled matrix protein 1 of influenza A virus.
Nature, 587, 2020
|
|
3NBI
 
 | | Crystal structure of human RMI1 N-terminus | | 分子名称: | RecQ-mediated genome instability protein 1 | | 著者 | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | 登録日 | 2010-06-03 | | 公開日 | 2010-09-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
3NBH
 
 | | Crystal structure of human RMI1C-RMI2 complex | | 分子名称: | RecQ-mediated genome instability protein 1, RecQ-mediated genome instability protein 2 | | 著者 | Wang, F, Yang, Y, Singh, T.R, Busygina, V, Guo, R, Wan, K, Wang, W, Sung, P, Meetei, A.R, Lei, M. | | 登録日 | 2010-06-03 | | 公開日 | 2010-09-22 | | 最終更新日 | 2017-11-08 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Crystal Structures of RMI1 and RMI2, Two OB-Fold Regulatory Subunits of the BLM Complex.
Structure, 18, 2010
|
|
7AQK
 
 | | Model of the actin filament Arp2/3 complex branch junction in cells | | 分子名称: | Actin, alpha skeletal muscle, ACTA1, ... | | 著者 | Faessler, F, Dimchev, G, Hodirnau, V.V, Wan, W, Schur, F.K.M. | | 登録日 | 2020-10-22 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Cryo-electron tomography structure of Arp2/3 complex in cells reveals new insights into the branch junction.
Nat Commun, 11, 2020
|
|
5O4W
 
 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | 分子名称: | Lysozyme C | | 著者 | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | 主引用文献 | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5O4X
 
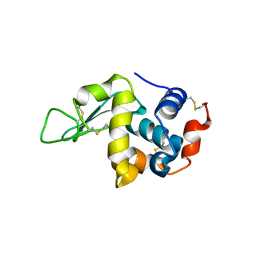 | | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal | | 分子名称: | Lysozyme C | | 著者 | Clabbers, M.T.B, van Genderen, E, Wan, W, Wiegers, E.L, Gruene, T, Abrahams, J.P. | | 登録日 | 2017-05-31 | | 公開日 | 2017-08-23 | | 最終更新日 | 2024-01-17 | | 実験手法 | ELECTRON CRYSTALLOGRAPHY (2.11 Å) | | 主引用文献 | Protein structure determination by electron diffraction using a single three-dimensional nanocrystal.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4JGV
 
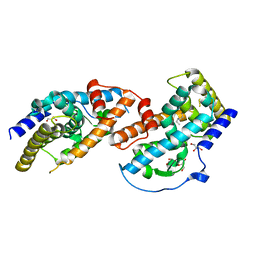 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with THPN | | 分子名称: | 1-(3,4,5-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | 著者 | Zhang, Q, Li, F, Li, A, Tian, X, Wan, W, Wan, Y, Chen, H, Xing, Y, Wu, Q, Lin, T. | | 登録日 | 2013-03-04 | | 公開日 | 2013-12-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.01 Å) | | 主引用文献 | Orphan nuclear receptor TR3 acts in autophagic cell death via mitochondrial signaling pathway.
Nat.Chem.Biol., 10, 2014
|
|
5L93
 
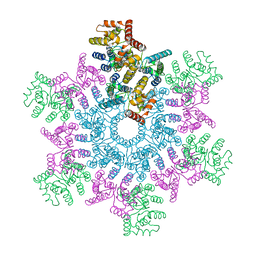 | | An atomic model of HIV-1 CA-SP1 reveals structures regulating assembly and maturation | | 分子名称: | Capsid protein p24 | | 著者 | Schur, F.K.M, Obr, M, Hagen, W.J.H, Wan, W, Arjen, J.J, Kirkpatrick, J.M, Sachse, C, Kraeusslich, H.-G, Briggs, J.A.G. | | 登録日 | 2016-06-09 | | 公開日 | 2016-07-13 | | 最終更新日 | 2024-05-15 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation.
Science, 353, 2016
|
|
7ELG
 
 | | LC3B modificated with a covalent probe | | 分子名称: | 2-methylidene-5-thiophen-2-yl-cyclohexane-1,3-dione, Microtubule-associated proteins 1A/1B light chain 3B, SULFATE ION | | 著者 | Fan, S, Wan, W. | | 登録日 | 2021-04-10 | | 公開日 | 2021-10-13 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.599 Å) | | 主引用文献 | Inhibition of Autophagy by a Small Molecule through Covalent Modification of the LC3 Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7FIV
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidA and CidBND1-ND2 from wPip(Tunis) | | 分子名称: | CidA_I gamma/2 protein, CidB_I b/2 protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIW
 
 | | Crystal structure of the complex formed by Wolbachia cytoplasmic incompatibility factors CidAwMel(ST) and CidBND1-ND2 from wPip(Pel) | | 分子名称: | ULP_PROTEASE domain-containing protein, bacteria factor 4,CidA I(Zeta/1) protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIU
 
 | | Crystal structure of the DUB domain of Wolbachia cytoplasmic incompatibility factor CidB from wMel | | 分子名称: | ULP_PROTEASE domain-containing protein | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.84 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
7FIT
 
 | | Crystal structure of Wolbachia cytoplasmic incompatibility factor CidA from wMel | | 分子名称: | bacteria factor 1 | | 著者 | Xiao, Y.J, Wang, W, Chen, X, Ji, X.Y, Yang, H.T. | | 登録日 | 2021-08-01 | | 公開日 | 2022-04-06 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Crystal Structures of Wolbachia CidA and CidB Reveal Determinants of Bacteria-induced Cytoplasmic Incompatibility and Rescue.
Nat Commun, 13, 2022
|
|
1JE9
 
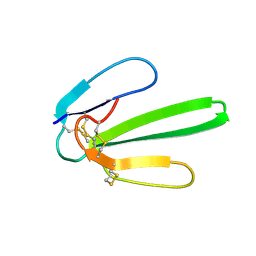 | | NMR SOLUTION STRUCTURE OF NT2 | | 分子名称: | SHORT NEUROTOXIN II | | 著者 | Cheng, Y, Wang, W, Wang, J. | | 登録日 | 2001-06-16 | | 公開日 | 2001-07-04 | | 最終更新日 | 2022-02-23 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure-function relationship of three neurotoxins from the venom of Naja kaouthia: a comparison between the NMR-derived structure of NT2 with its homologues, NT1 and NT3
BIOCHIM.BIOPHYS.ACTA, 1594, 2002
|
|
6SKT
 
 | |
4LVA
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | 分子名称: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | 著者 | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | 登録日 | 2013-07-26 | | 公開日 | 2013-09-25 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
