7DY7
 
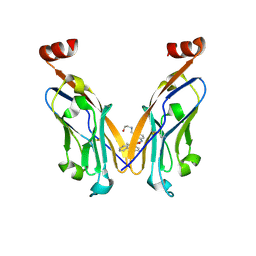 | | Discovery of Novel Small-molecule Inhibitors of PD-1/PD-L1 Axis that Promotes PD-L1 Internalization and Degradation | | Descriptor: | 2-[[3-[[5-(2-methyl-3-phenyl-phenyl)-1,3,4-oxadiazol-2-yl]amino]phenyl]methylamino]ethanol, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Wang, T.Y, Lu, M.L, Jiang, S, Xiao, Y.B. | | Deposit date: | 2021-01-20 | | Release date: | 2022-01-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Discovery of Small-Molecule Inhibitors of the PD-1/PD-L1 Axis That Promote PD-L1 Internalization and Degradation.
J.Med.Chem., 65, 2022
|
|
4XX5
 
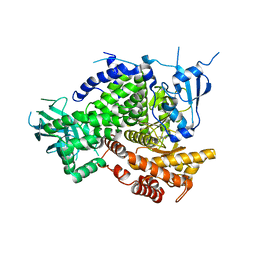 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(5-methoxypyridin-3-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-01-29 | | Release date: | 2015-08-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
1FKT
 
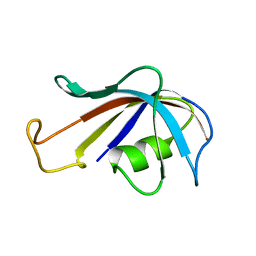 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
4XZ4
 
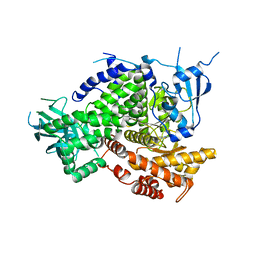 | | Structure of PI3K gamma in complex with an inhibitor | | Descriptor: | N-[5-(6-methoxypyrazin-2-yl)-4,5,6,7-tetrahydro[1,3]thiazolo[5,4-c]pyridin-2-yl]acetamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Collier, P.N, Messersmith, D, Le Tiran, A, Bandarage, U.K, Boucher, C, Come, J, Cottrell, K.M, Damagnez, V, Doran, J.D, Griffith, J.P, Khare-Pandit, S, Krueger, E.B, Ledeboer, M.W, Ledford, B, Liao, Y, Mahajan, S, Moody, C.S, Wang, T, Xu, J, Aronov, A.M. | | Deposit date: | 2015-02-03 | | Release date: | 2016-02-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PI3K gamma in complex with an inhibitor
To Be Published
|
|
1FKR
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
1FKS
 
 | | SOLUTION STRUCTURE OF FKBP, A ROTAMASE ENZYME AND RECEPTOR FOR FK506 AND RAPAMYCIN | | Descriptor: | FK506 AND RAPAMYCIN-BINDING PROTEIN | | Authors: | Michnick, S.W, Rosen, M.K, Wandless, T.J, Karplus, M, Schreiber, S.L. | | Deposit date: | 1992-03-05 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin.
Science, 252, 1991
|
|
3KCK
 
 | | A Novel Chemotype of Kinase Inhibitors | | Descriptor: | 3-chloro-4-(4H-3,4,7-triazadibenzo[cd,f]azulen-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Zuccola, H.J, Wang, T, Ledeboer, M.W. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A novel chemotype of kinase inhibitors: Discovery of 3,4-ring fused 7-azaindoles and deazapurines as potent JAK2 inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
1YHN
 
 | | Structure basis of RILP recruitment by Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab interacting lysosomal protein, ... | | Authors: | Wu, M, Wang, T, Hong, W, Song, H. | | Deposit date: | 2005-01-10 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for recruitment of RILP by small GTPase Rab7.
Embo J., 24, 2005
|
|
5UGK
 
 | | Zinc-Binding Structure of a Catalytic Amyloid from Solid-State NMR Spectroscopy | | Descriptor: | ILE-HIS-VAL-HIS-LEU-GLN-ILE, ZINC ION | | Authors: | Lee, M, Wang, T, Makhlynets, O.V, Wu, Y, Polizzi, N, Wu, H, Gosavi, P.M, Korendovych, I.V, DeGrado, W.F, Hong, M. | | Deposit date: | 2017-01-09 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Zinc-binding structure of a catalytic amyloid from solid-state NMR.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5F1M
 
 | |
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
4DHI
 
 | | Structure of C. elegans OTUB1 bound to human UBC13 | | Descriptor: | Ubiquitin thioesterase otubain-like, Ubiquitin-conjugating enzyme E2 N | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-27 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
7MEY
 
 | | Structure of yeast Ubr1 in complex with Ubc2 and monoubiquitinated N-degron | | Descriptor: | 2-(ethylamino)ethane-1-thiol, E3 ubiquitin-protein ligase UBR1, Monoubiquitinated N-degron, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2021-12-22 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
7MEX
 
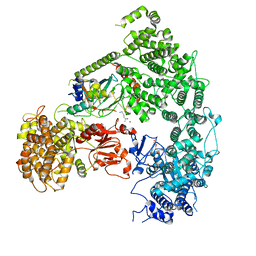 | | Structure of yeast Ubr1 in complex with Ubc2 and N-degron | | Descriptor: | E3 ubiquitin-protein ligase UBR1, N-degron, Ubiquitin, ... | | Authors: | Pan, M, Zheng, Q, Wang, T, Liang, L, Yu, Y, Liu, L, Zhao, M. | | Deposit date: | 2021-04-08 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into Ubr1-mediated N-degron polyubiquitination.
Nature, 600, 2021
|
|
3UBI
 
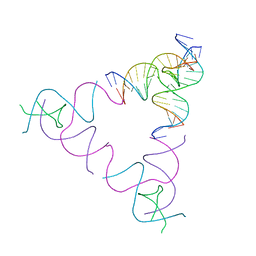 | | The Absence of Tertiary Interactions in a Self-Assembled DNA Crystal Structure | | Descriptor: | DNA (5'-D(*GP*AP*GP*CP*AP*GP*CP*CP*CP*GP*TP*AP*CP*TP*CP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*AP*GP*GP*CP*TP*GP*C)-3'), DNA (5'-D(P*CP*CP*GP*AP*GP*TP*AP*CP*GP*AP*CP*GP*AP*CP*AP*AP*G)-3'), ... | | Authors: | Nguyen, N, Birktoft, J.J, Sha, R, Wang, T, Zheng, J, Constantinou, P.E, Ginell, S.L, Chen, Y, Mao, C, Seeman, N.C. | | Deposit date: | 2011-10-24 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (6.8046 Å) | | Cite: | The absence of tertiary interactions in a self-assembled DNA crystal structure.
J.Mol.Recognit., 25, 2012
|
|
6JK8
 
 | | Cryo-EM structure of the full-length human IGF-1R in complex with insulin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Zhang, X, Yu, D, Wang, T. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Visualization of Ligand-Bound Ectodomain Assembly in the Full-Length Human IGF-1 Receptor by Cryo-EM Single-Particle Analysis.
Structure, 28, 2020
|
|
7BWA
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 2 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
3M9H
 
 | |
8AGQ
 
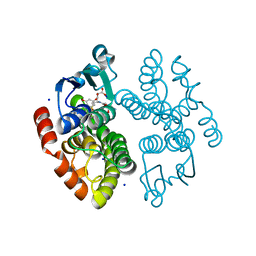 | | Crystal structure of anthocyanin-related GSTF8 from Populus trichocarpa in complex with (-)-catechin and glutathione | | Descriptor: | (2~{S},3~{R})-2-[3,4-bis(oxidanyl)phenyl]-3,4-dihydro-2~{H}-chromene-3,5,7-triol, GLUTATHIONE, Glutathione transferase, ... | | Authors: | Eichenberger, M, Hueppi, S, Schwander, T, Mittl, P, Buller, M.R. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.093 Å) | | Cite: | The catalytic role of glutathione transferases in heterologous anthocyanin biosynthesis.
Nat Catal, 6, 2023
|
|
4EFB
 
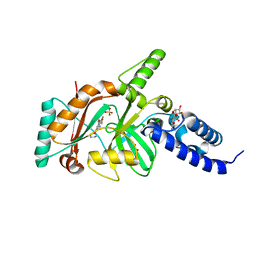 | | Crystal structure of DNA ligase | | Descriptor: | 4-amino-2-(cyclopentyloxy)-6-{[(1R,2S)-2-hydroxycyclopentyl]oxy}pyrimidine-5-carboxamide, BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of DNA ligase
To be Published
|
|
7BW7
 
 | | Cryo-EM Structure for the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin. | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-13 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
7BW8
 
 | | Cryo-EM Structure for the Insulin Binding Region in the Ectodomain of the Full-length Human Insulin Receptor in Complex with 1 Insulin | | Descriptor: | Insulin fusion, Insulin receptor | | Authors: | Yu, D, Zhang, X, Sun, J, Li, X, Wu, Z, Han, X, Fan, C, Ma, Y, Ouyang, Q, Wang, T. | | Deposit date: | 2020-04-14 | | Release date: | 2021-04-14 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Insulin Binding Induced the Ectodomain Conformational Dynamics in the Full-length Human Insulin Receptor
To Be Published
|
|
1QHI
 
 | | HERPES SIMPLEX VIRUS TYPE-I THYMIDINE KINASE COMPLEXED WITH A NOVEL NON-SUBSTRATE INHIBITOR, 9-(4-HYDROXYBUTYL)-N2-PHENYLGUANINE | | Descriptor: | 9-(4-HYDROXYBUTYL)-N2-PHENYLGUANINE, PROTEIN (THYMIDINE KINASE), SULFATE ION | | Authors: | Bennett, M.S, Wien, F, Champness, J.N, Batuwangala, T, Rutherford, T, Summers, W.C, Sun, H, Wright, G, Sanderson, M.R. | | Deposit date: | 1999-05-12 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure to 1.9 A resolution of a complex with herpes simplex virus type-1 thymidine kinase of a novel, non-substrate inhibitor: X-ray crystallographic comparison with binding of aciclovir.
FEBS Lett., 443, 1999
|
|
4EFE
 
 | | crystal structure of DNA ligase | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DNA ligase, SULFATE ION, ... | | Authors: | Wei, Y, Wang, T, Charifson, P, Xu, W. | | Deposit date: | 2012-03-29 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure of DNA ligase
To be Published
|
|
2LRW
 
 | | Solution structure of a ubiquitin-like protein from Trypanosoma brucei | | Descriptor: | Ubiquitin, putative | | Authors: | Wang, R, Wang, T, Liao, S, Zhang, J, Tu, X. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a ubiqutin-like protein from Trypanosoma bucei
To be Published
|
|
