7CXM
 
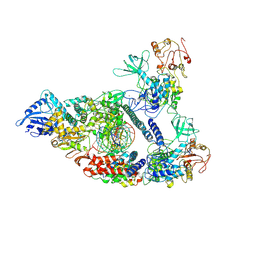 | | Architecture of a SARS-CoV-2 mini replication and transcription complex | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L, Zhang, Y, Ge, J, Zheng, L, Gao, Y, Wang, T, Jia, Z, Wang, H, Huang, Y, Li, M, Wang, Q, Rao, Z, Lou, Z. | | Deposit date: | 2020-09-02 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Architecture of a SARS-CoV-2 mini replication and transcription complex.
Nat Commun, 11, 2020
|
|
5Z23
 
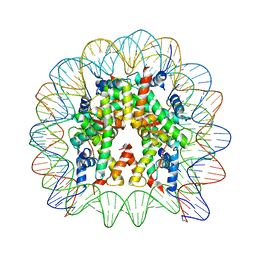 | | Crystal structure of the nucleosome containing a chimeric histone H3/CENP-A CATD | | Descriptor: | DNA (146-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Arimura, Y, Tachiwana, H, Takagi, H. | | Deposit date: | 2017-12-28 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The CENP-A centromere targeting domain facilitates H4K20 monomethylation in the nucleosome by structural polymorphism.
Nat Commun, 10, 2019
|
|
5B24
 
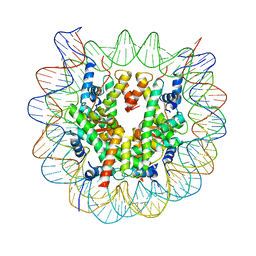 | | The crystal structure of the nucleosome containing cyclobutane pyrimidine dimer | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Horikoshi, N, Tachiwana, H, Kagawa, W, Osakabe, A, Kurumizaka, H. | | Deposit date: | 2015-12-31 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Crystal structure of the nucleosome containing ultraviolet light-induced cyclobutane pyrimidine dimer
Biochem.Biophys.Res.Commun., 471, 2016
|
|
3NV6
 
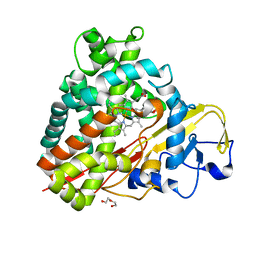 | | Crystal Structure of Camphor-Bound CYP101D2 | | Descriptor: | CAMPHOR, Cytochrome P450, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Zhou, W.H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of CYP101D2 unveils a potential path for substrate entry into the active site
Biochem.J., 433, 2011
|
|
3TDL
 
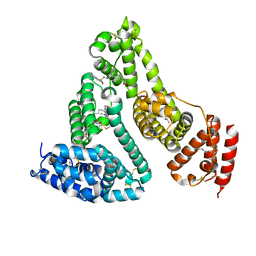 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
3M03
 
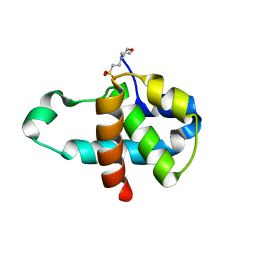 | | Crystal structure of human Orc6 fragment | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Origin recognition complex subunit 6, SULFATE ION | | Authors: | Liu, S.X, Wu, L.J, Sun, J, Wang, H.F, Liu, Y.F. | | Deposit date: | 2010-03-02 | | Release date: | 2011-03-09 | | Last modified: | 2014-03-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis of human Orc6 protein reveals a homology with transcription factor TFIIB
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4NFB
 
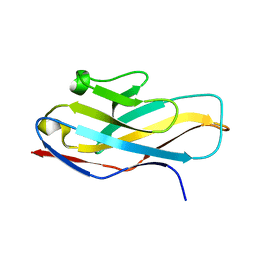 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor alpha | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NFD
 
 | | Structure of PILR L108W mutant in complex with sialic acid | | Descriptor: | N-acetyl-alpha-neuraminic acid, Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KAC
 
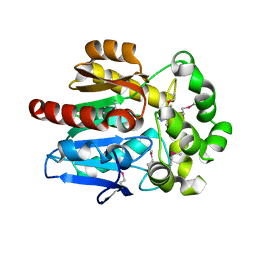 | | X-Ray Structure of the complex HaloTag2 with HALTS. Northeast Structural Genomics Consortium (NESG) Target OR150. | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, AMMONIUM ION, Haloalkane dehalogenase, ... | | Authors: | Kuzin, A, Lew, S, Neklesa, T.K, Noblin, D, Seetharaman, J, Maglaqui, M, Xiao, R, Kohan, E, Wang, H, Everett, J.K, Acton, T.B, Kornhaber, G, Montelione, G.T, Crews, C.M, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-04-22 | | Release date: | 2013-06-05 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.223 Å) | | Cite: | Northeast Structural Genomics Consortium Target OR150
To be Published
|
|
4NFC
 
 | | Structure of paired immunoglobulin-like type 2 receptor (PILR ) | | Descriptor: | Paired immunoglobulin-like type 2 receptor beta | | Authors: | Lu, Q, Lu, G, Qi, J, Li, Y, Zhang, Y, Wang, H, Fan, Z, Yan, J, Gao, G.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PILR alpha and PILR beta have a siglec fold and provide the basis of binding to sialic acid
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3LXI
 
 | | Crystal Structure of Camphor-Bound CYP101D1 | | Descriptor: | CAMPHOR, Cytochrome P450, PHOSPHATE ION, ... | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
2QC3
 
 | | Crystal structure of MCAT from Mycobacterium tuberculosis | | Descriptor: | ACETIC ACID, Malonyl CoA-acyl carrier protein transacylase | | Authors: | Li, Z, Huang, Y, Ge, J, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2007-06-19 | | Release date: | 2007-08-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of MCAT from Mycobacterium tuberculosis Reveals Three New Catalytic Models.
J.Mol.Biol., 371, 2007
|
|
2JVM
 
 | | Solution NMR structure of Rhodobacter sphaeroides protein RHOS4_26430. Northeast Structural Genomics Consortium target RhR95 | | Descriptor: | Uncharacterized protein | | Authors: | Eletsky, A, Sukumaran, D, Zhang, Q, Parish, D, Xu, D, Wang, H, Janjua, H, Owens, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-21 | | Release date: | 2007-10-16 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Rhodobacter sphaeroides protein RHOS4_26430.
To be Published
|
|
2LFC
 
 | | Solution NMR Structure of Fumarate reductase flavoprotein subunit from Lactobacillus plantarum, Northeast Structural Genomics Consortium Target LpR145J | | Descriptor: | Fumarate reductase, flavoprotein subunit | | Authors: | Liu, G, Tang, Y, Xiao, R, Janjua, H, Ciccosanti, C, Wang, H, Acton, T, Everett, J.K, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-29 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target LpR145J
To be Published
|
|
2L9R
 
 | | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A | | Descriptor: | Homeobox protein Nkx-3.1 | | Authors: | Liu, G, Xiao, R, Lee, H.-W, Hamilton, K, Ciccosanti, C, Wang, H.B, Acton, T.B, Everett, J.K, Huang, Y.J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Homeobox domain of Homeobox protein Nkx-3.1 from homo sapiens, Northeast Structural Genomics Consortium Target HR6470A
To be Published
|
|
2MXD
 
 | | Solution structure of VPg of porcine sapovirus | | Descriptor: | Viral protein genome-linked | | Authors: | Kim, J, Hwang, H, Min, H, Yun, H, Cho, K, Pelton, J.G, Wemmer, D.E, Lee, C. | | Deposit date: | 2014-12-24 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Biochem.Biophys.Res.Commun., 459, 2015
|
|
2LEK
 
 | | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris refined with NH RDCs. Northeast Structural Genomics Consortium target RpR325 | | Descriptor: | Putative thiamin biosynthesis ThiS | | Authors: | Ramelot, T.A, Cort, J.R, Lee, H, Wang, H, Ciccosanti, C, Jiang, M, Nair, R, Rost, B, Acton, T.B, Xiao, R, Swapna, G, Everett, J.K, Prestegard, J.H, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of a Thiamine Biosynthesis (ThiS) Protein RPA3574 from Rhodopseudomonas palustris. Northeast Structural Genomics Consortium target RpR325
To be Published
|
|
7C8U
 
 | | The crystal structure of COVID-19 main protease in complex with GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase | | Authors: | Luan, X, Shang, W, Wang, Y, Yin, W, Jiang, Y, Feng, S, Wang, Y, Liu, M, Zhou, R, Zhang, Z, Wang, F, Cheng, W, Gao, M, Wang, H, Wu, W, Tian, R, Tian, Z, Jin, Y, Jiang, H.W, Zhang, L, Xu, H.E, Zhang, S. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The crystal structure of COVID-19 main protease in complex with GC376
To Be Published
|
|
3LBA
 
 | |
2Z8O
 
 | | Structural basis for the catalytic mechanism of phosphothreonine lyase | | Descriptor: | 27.5 kDa virulence protein, L(+)-TARTARIC ACID | | Authors: | Chen, L, Wang, H, Gu, L, Huang, N, Zhou, J.M, Chai, J. | | Deposit date: | 2007-09-07 | | Release date: | 2007-12-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the catalytic mechanism of phosphothreonine lyase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3LXD
 
 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | Descriptor: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | Deposit date: | 2010-02-25 | | Release date: | 2010-06-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
4C5Z
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
7DGB
 
 | |
7DGH
 
 | |
7DGF
 
 | |
