6J6I
 
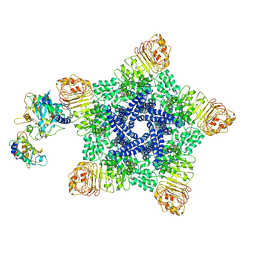 | | Reconstitution and structure of a plant NLR resistosome conferring immunity | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, ... | | 著者 | Wang, J.Z, Wang, J, Hu, M.J, Wang, H.W, Zhou, J.M, Chai, J.J. | | 登録日 | 2019-01-15 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-04-12 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Reconstitution and structure of a plant NLR resistosome conferring immunity.
Science, 364, 2019
|
|
3UDP
 
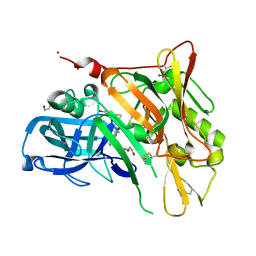 | | Crystal Structure of BACE with Compound 12 | | 分子名称: | (4S)-6-bromo-3,4-dihydro-2H-thiochromen-4-yl (3S,5'R)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxylate, 1,2-ETHANEDIOL, Beta-secretase 1, ... | | 著者 | Efremov, I.V, Vajdos, F.F, Borzilleri, K, Capetta, S, Dorff, P, Dutra, J, Mansour, M, Oborski, C, O'Connell, T, O'Sullivan, T.J, Pandit, J, Wang, H, Withka, J. | | 登録日 | 2011-10-28 | | 公開日 | 2012-04-18 | | 最終更新日 | 2018-04-04 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Discovery and optimization of a novel spiropyrrolidine inhibitor of {beta}-secretase (BACE1) through fragment-based drug design.
J.Med.Chem., 55, 2012
|
|
6LYV
 
 | |
5XMI
 
 | | Cryo-EM Structure of the ATP-bound VPS4 mutant-E233Q hexamer (masked) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Vacuolar protein sorting-associated protein 4 | | 著者 | Sun, S, Li, L, Yang, F, Wang, X, Fan, F, Li, X, Wang, H, Sui, S. | | 登録日 | 2017-05-15 | | 公開日 | 2017-08-09 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Cryo-EM structures of the ATP-bound Vps4(E233Q) hexamer and its complex with Vta1 at near-atomic resolution
Nat Commun, 8, 2017
|
|
6J5U
 
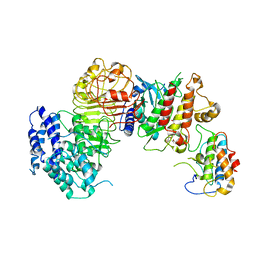 | | Ligand-triggered allosteric ADP release primes a plant NLR complex | | 分子名称: | Disease resistance RPP13-like protein 4, Probable serine/threonine-protein kinase PBL2, Protein kinase superfamily protein, ... | | 著者 | Wang, J.Z, Wang, J, Meijuan, H, Wang, H.W, Zhou, J.M, Chai, J.J. | | 登録日 | 2019-01-12 | | 公開日 | 2019-04-03 | | 最終更新日 | 2023-04-05 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Ligand-triggered allosteric ADP release primes a plant NLR complex.
Science, 364, 2019
|
|
6J6K
 
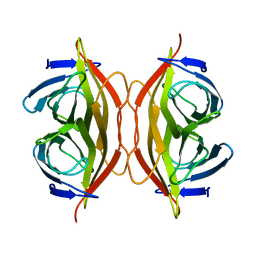 | | Apo-state streptavidin | | 分子名称: | Streptavidin | | 著者 | Fan, X, Wang, J, Lei, J.L, Wang, H.W. | | 登録日 | 2019-01-15 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Single particle cryo-EM reconstruction of 52 kDa streptavidin at 3.2 Angstrom resolution.
Nat Commun, 10, 2019
|
|
3LBA
 
 | |
3LXD
 
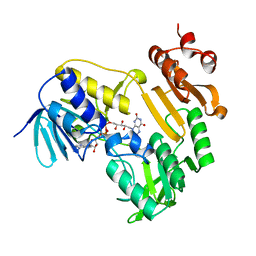 | | Crystal Structure of Ferredoxin Reductase ArR from Novosphingobium aromaticivorans | | 分子名称: | FAD-dependent pyridine nucleotide-disulphide oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | 著者 | Yang, W, Bell, S.G, Wang, H, Bartlam, M, Wong, L.L, Rao, Z. | | 登録日 | 2010-02-25 | | 公開日 | 2010-06-23 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444
J.Biol.Chem., 285, 2010
|
|
5YFP
 
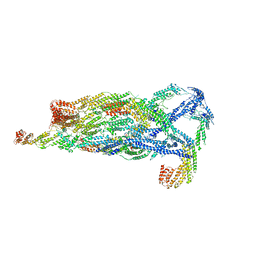 | | Cryo-EM Structure of the Exocyst Complex | | 分子名称: | Exocyst complex component EXO70, Exocyst complex component EXO84, Exocyst complex component SEC10, ... | | 著者 | Mei, K, Li, Y, Wang, S, Shao, G, Wang, J, Ding, Y, Luo, G, Yue, P, Liu, J.J, Wang, X, Dong, M.Q, Guo, W, Wang, H.W. | | 登録日 | 2017-09-21 | | 公開日 | 2018-01-31 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Cryo-EM structure of the exocyst complex
Nat. Struct. Mol. Biol., 25, 2018
|
|
4C60
 
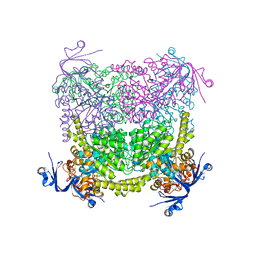 | | Crystal structure of A. niger ochratoxinase | | 分子名称: | OCHRATOXINASE | | 著者 | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | 登録日 | 2013-09-17 | | 公開日 | 2014-07-02 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
8HLL
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 1) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Wei, H, Guo, M, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLN
 
 | | Crystal structure of p53/BCL2 fusion complex(complex3) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wei, H, Wang, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.354 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
8HLM
 
 | | Crystal structure of p53/BCL2 fusion complex (complex 2) | | 分子名称: | Apoptosis regulator Bcl-2, Cellular tumor antigen p53, ZINC ION | | 著者 | Guo, M, Wang, H, Wei, H, Chen, Y. | | 登録日 | 2022-11-30 | | 公開日 | 2023-07-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.522 Å) | | 主引用文献 | Structures of p53/BCL-2 complex suggest a mechanism for p53 to antagonize BCL-2 activity.
Nat Commun, 14, 2023
|
|
5GW4
 
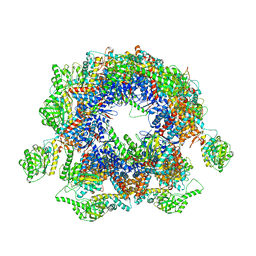 | | Structure of Yeast NPP-TRiC | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Zang, Y, Jin, M, Wang, H, Cong, Y. | | 登録日 | 2016-09-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.7 Å) | | 主引用文献 | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
7WC5
 
 | | Crystal structure of serotonin 2A receptor in complex with psilocin | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 3-[2-(dimethylamino)ethyl]-1~{H}-indol-4-ol, 5-hydroxytryptamine receptor 2A, ... | | 著者 | Cao, D, Yu, J, Wang, H, Luo, Z, Liu, X, He, L, Qi, J, Fan, L, Tang, L, Chen, Z, Li, J, Cheng, J, Wang, S. | | 登録日 | 2021-12-18 | | 公開日 | 2022-01-26 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure-based discovery of nonhallucinogenic psychedelic analogs.
Science, 375, 2022
|
|
2HD1
 
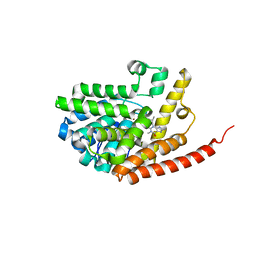 | | Crystal structure of PDE9 in complex with IBMX | | 分子名称: | 3-ISOBUTYL-1-METHYLXANTHINE, MAGNESIUM ION, Phosphodiesterase 9A, ... | | 著者 | Huai, Q, Wang, H, Zhang, W, Colman, R.W, Robinson, H, Ke, H. | | 登録日 | 2006-06-19 | | 公開日 | 2006-06-27 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.23 Å) | | 主引用文献 | Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor IBMX binding
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6J15
 
 | | Complex structure of GY-5 Fab and PD-1 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GY-5 heavy chain Fab, ... | | 著者 | Chen, D, Tan, S, Zhang, H, Wang, H, Chai, Y, Qi, J, Yan, J, Gao, G.F. | | 登録日 | 2018-12-27 | | 公開日 | 2019-11-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | The FG Loop of PD-1 Serves as a "Hotspot" for Therapeutic Monoclonal Antibodies in Tumor Immune Checkpoint Therapy.
Iscience, 14, 2019
|
|
5GW5
 
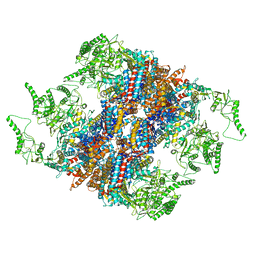 | | Structure of TRiC-AMP-PNP | | 分子名称: | T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, T-complex protein 1 subunit delta, ... | | 著者 | Zang, Y, Jin, M, Wang, H, Cong, Y. | | 登録日 | 2016-09-08 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Staggered ATP binding mechanism of eukaryotic chaperonin TRiC (CCT) revealed through high-resolution cryo-EM.
Nat. Struct. Mol. Biol., 23, 2016
|
|
8HRU
 
 | |
8HRK
 
 | |
8HRN
 
 | | Cryo-EM structure of ACE2 | | 分子名称: | Angiotensin-converting enzyme 2 | | 著者 | Xu, J, Liu, N, Wang, H.W. | | 登録日 | 2022-12-15 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | 分子名称: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | 著者 | Xu, J, Meng, F, Liu, N, Wang, H.W. | | 登録日 | 2022-12-15 | | 公開日 | 2023-12-20 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
5YIJ
 
 | | Structure of a Legionella effector with substrates | | 分子名称: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, SdeA, Ubiquitin | | 著者 | Feng, Y, Mu, Y, Wang, H. | | 登録日 | 2017-10-05 | | 公開日 | 2018-05-30 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.18 Å) | | 主引用文献 | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
5YYS
 
 | | Cryo-EM structure of L-fucokinase, GDP-fucose pyrophosphorylase (FKP)in Bacteroides fragilis | | 分子名称: | L-fucokinase, L-fucose-1-P guanylyltransferase | | 著者 | Wang, J, Hu, H, Liu, Y, Zhou, Q, Wu, P, Yan, N, Wang, H.W, Wu, J.W, Sun, L. | | 登録日 | 2017-12-11 | | 公開日 | 2018-12-12 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4.2 Å) | | 主引用文献 | Cryo-EM structure of L-fucokinase/GDP-fucose pyrophosphorylase (FKP) in Bacteroides fragilis.
Protein Cell, 10, 2019
|
|
6JPD
 
 | | Mouse receptor-interacting protein kinase 3 (RIP3) amyloid structure by solid-state NMR | | 分子名称: | Receptor-interacting serine/threonine-protein kinase 3 | | 著者 | Wu, X.L, Hu, H, Zhang, J, Dong, X.Q, Wang, J, Schwieters, C, Wang, H.Y, Lu, J.X. | | 登録日 | 2019-03-26 | | 公開日 | 2020-10-28 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLID-STATE NMR | | 主引用文献 | The amyloid structure of mouse RIPK3 (receptor interacting protein kinase 3) in cell necroptosis.
Nat Commun, 12, 2021
|
|
