7L7V
 
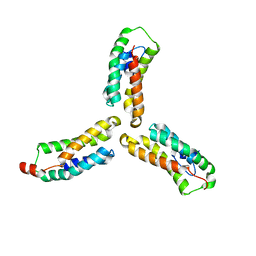 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E/R99E/K100E/R103E/K106E/K110E mutant | | Descriptor: | Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
7L7W
 
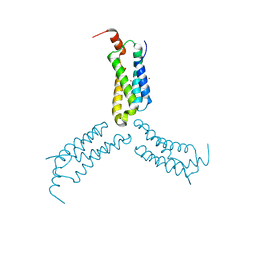 | | Crystal structure of Arabidopsis NRG1.1 CC-R domain K94E/K96E mutant | | Descriptor: | NICKEL (II) ION, Probable disease resistance protein At5g66900 | | Authors: | Walton, W.G, Wan, L, Lietzan, A.D, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-12-30 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Plant "helper" immune receptors are Ca 2+ -permeable nonselective cation channels.
Science, 373, 2021
|
|
6NCX
 
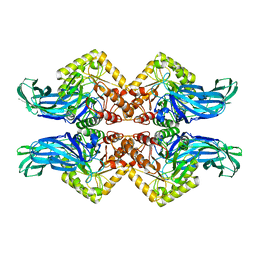 | |
6P2B
 
 | | Tethered PXR-LBD/SRC-1p bound to Garcinoic Acid | | Descriptor: | (2Z,6E,10E)-13-[(2R)-6-hydroxy-2,8-dimethyl-3,4-dihydro-2H-1-benzopyran-2-yl]-2,6,10-trimethyltrideca-2,6,10-trienoic acid, DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2019-05-21 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Garcinoic Acid Is a Natural and Selective Agonist of Pregnane X Receptor.
J.Med.Chem., 63, 2020
|
|
5UJ6
 
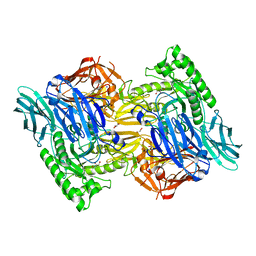 | |
6D1P
 
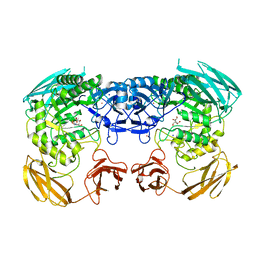 | | Apo structure of Bacteroides uniformis beta-glucuronidase 3 | | Descriptor: | GLYCEROL, Glycosyl hydrolases family 2, sugar binding domain protein, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-12 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D41
 
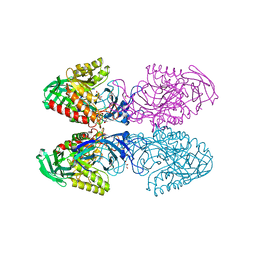 | | Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-17 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D50
 
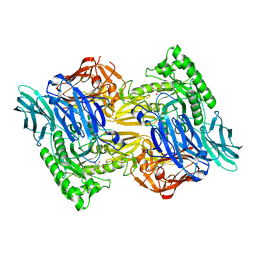 | | Bacteroides uniforms beta-glucuronidase 2 bound to D-glucaro-1,5-lactone | | Descriptor: | (2S,3S,4S,5R)-3,4,5-trihydroxy-6-oxo-oxane-2-carboxylic acid, CALCIUM ION, Glycosyl hydrolases family 2, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-19 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D89
 
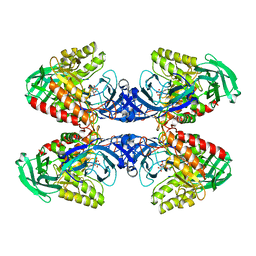 | |
5I7J
 
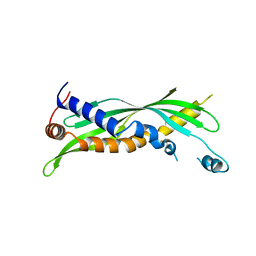 | |
5I7K
 
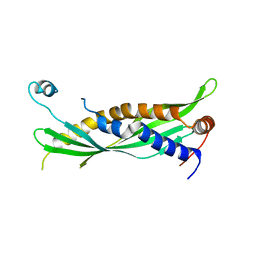 | | Crystal Structure of Human SPLUNC1 Dolphin Mutant D1 (G58A, S61A, G62E, G63D, G66D, I67T) | | Descriptor: | BPI fold-containing family A member 1 | | Authors: | Walton, W.G, Redinbo, M.R. | | Deposit date: | 2016-02-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.552 Å) | | Cite: | Structural Features Essential to the Antimicrobial Functions of Human SPLUNC1.
Biochemistry, 55, 2016
|
|
5I7L
 
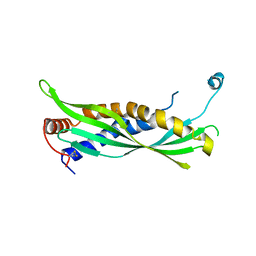 | |
6NCZ
 
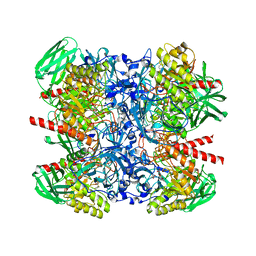 | |
6NCW
 
 | |
6NCY
 
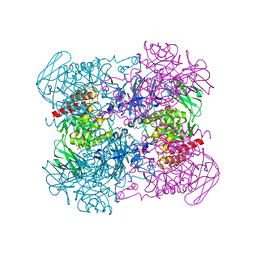 | | Crystal structure of hybrid beta-glucuronidase/beta-galacturonidase from Fusicatenibacter saccharivorans | | Descriptor: | Beta-glucuronidase, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-12-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Selecting a Single Stereocenter: The Molecular Nuances That Differentiate beta-Hexuronidases in the Human Gut Microbiome.
Biochemistry, 58, 2019
|
|
8DTQ
 
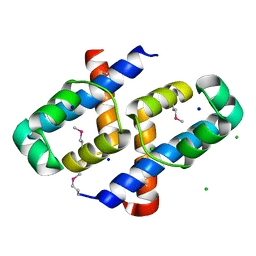 | | Crystal Structure of Staphylococcus aureus pSK41 Cop | | Descriptor: | CHLORIDE ION, Helix-turn-helix domain, SODIUM ION | | Authors: | Walton, W.G, Eakes, T.C, Redinbo, M.R, McLaughlin, K.J. | | Deposit date: | 2022-07-26 | | Release date: | 2023-08-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | pSK41/pGO1-family conjugative plasmids of Staphylococcus aureus encode a cryptic repressor of replication.
Plasmid, 128, 2023
|
|
6D1N
 
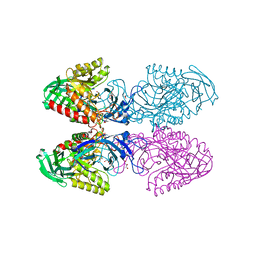 | |
6D6W
 
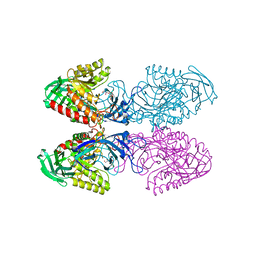 | | Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-23 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D8G
 
 | |
6D7F
 
 | | Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide | | Descriptor: | Beta-galactosidase/beta-glucuronidase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Walton, W.G, Pellock, S.J, Redinbo, M.R. | | Deposit date: | 2018-04-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three structurally and functionally distinct beta-glucuronidases from the human gut microbeBacteroides uniformis.
J. Biol. Chem., 293, 2018
|
|
6D8K
 
 | |
7KFO
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 acetic acid | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KFQ
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to 1H-Indole-3-Carboxylic Acid | | Descriptor: | 1H-INDOLE-3-CARBOXYLIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KFS
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Benzoic Acid | | Descriptor: | BENZOIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-14 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
7KH3
 
 | | Crystal structure of the MarR family transcriptional regulator from Variovorax paradoxus bound to Indole 3 propionic acid | | Descriptor: | INDOLYLPROPIONIC ACID, SULFATE ION, Transcriptional regulator, ... | | Authors: | Walton, W.G, Redinbo, M.R, Dangl, J.L. | | Deposit date: | 2020-10-19 | | Release date: | 2021-12-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse MarR bacterial regulators of auxin catabolism in the plant microbiome.
Nat Microbiol, 7, 2022
|
|
