6P7E
 
 | | Structure of T7 DNA Polymerase Bound to a Primer/Template DNA and a Peptide that Mimics the C-terminal Tail of the Primase-Helicase | | Descriptor: | ASP-THR-ASP-PHE peptide, DNA (25-MER), DNA (5'-D(P*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*CP*TP*TP*GP*CP*CP*GP*GP*TP*GP*A)-3'), ... | | Authors: | Foster, B.M, Rosenberg, D, Salvo, H, Stephens, K.L, Bintz, B.J, Hammel, M, Ellenberger, T, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2019-06-05 | | Release date: | 2020-03-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Combined Solution and Crystal Methods Reveal the Electrostatic Tethers That Provide a Flexible Platform for Replication Activities in the Bacteriophage T7 Replisome.
Biochemistry, 58, 2019
|
|
7R6R
 
 | | Crystal Structure of a Mycobacteriophage Cluster A2 Immunity Repressor:DNA Complex | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*TP*TP*GP*AP*CP*AP*GP*CP*CP*AP*CP*CP*GP*AP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*CP*GP*GP*TP*GP*GP*CP*TP*GP*TP*CP*AP*AP*GP*CP*GP*GP*G)-3'), Immunity repressor | | Authors: | McGinnis, R.J, Brambley, C.A, Stamey, B, Green, W.C, Gragg, K.N, Cafferty, E.R, Terwilliger, T.C, Hammel, M, Hollis, T.J, Miller, J.M, Gainey, M.D, Wallen, J.R. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | A monomeric mycobacteriophage immunity repressor utilizes two domains to recognize an asymmetric DNA sequence.
Nat Commun, 13, 2022
|
|
6VE6
 
 | | A structural characterization of poly(aspartic acid) hydrolase-1 from Sphingomonas sp. KT-1. | | Descriptor: | Poly(Aspartic acid) hydrolase-1 | | Authors: | Bolay, A.L, Salvo, H, Brambley, C.A, Yared, T.J, Miller, J.M, Wallen, J.R, Weiland, M.H. | | Deposit date: | 2019-12-28 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.446 Å) | | Cite: | Structural Characterization of Sphingomonas sp. KT-1 PahZ1-Catalyzed Biodegradation of Thermally Synthesized Poly(aspartic acid)
Acs Sustain Chem Eng, 2020
|
|
3LRI
 
 | | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I | | Descriptor: | PROTEIN (INSULIN-LIKE GROWTH FACTOR I) | | Authors: | Laajoki, L.G, Francis, G.L, Wallace, J.C, Carver, J.A, Keniry, M.A. | | Deposit date: | 1999-04-13 | | Release date: | 2000-05-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of long-[Arg(3)]insulin-like growth factor-I
J.Biol.Chem., 275, 2000
|
|
5W0M
 
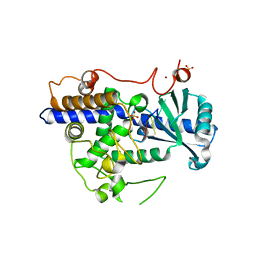 | | Structure of human TUT7 catalytic module (CM) in complex with U5 RNA | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0B
 
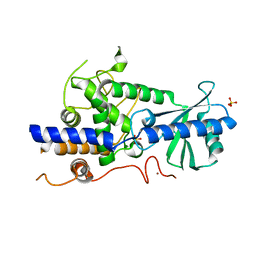 | | Structure of human TUT7 catalytic module (CM) | | Descriptor: | IODIDE ION, SULFATE ION, Terminal uridylyltransferase 7, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-30 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5W0O
 
 | |
5W0N
 
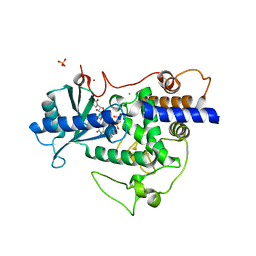 | | Structure of human TUT7 catalytic module (CM) in complex with UMPNPP and U2 RNA | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Faehnle, C.R, Walleshauser, J, Joshua-Tor, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1IGL
 
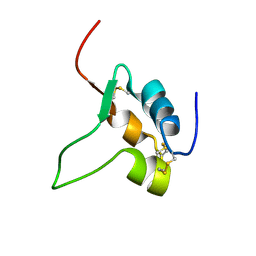 | | SOLUTION STRUCTURE OF HUMAN INSULIN-LIKE GROWTH FACTOR II RELATIONSHIP TO RECEPTOR AND BINDING PROTEIN INTERACTIONS | | Descriptor: | INSULIN-LIKE GROWTH FACTOR II | | Authors: | Torres, A.M, Forbes, B.E, Aplin, S.E, Wallace, J.C, Francis, G.L, Norton, R.S. | | Deposit date: | 1994-12-29 | | Release date: | 1995-02-14 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions.
J.Mol.Biol., 248, 1995
|
|
2BDO
 
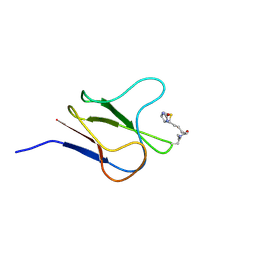 | | SOLUTION STRUCTURE OF HOLO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | BIOTIN, PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-03 | | Release date: | 1999-04-27 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
5DXU
 
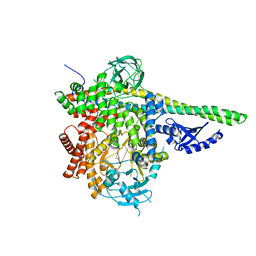 | | p110delta/p85alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DXH
 
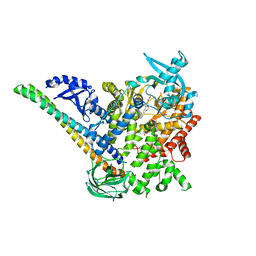 | | p110alpha/p85alpha with compound 5 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform, methyl {2-[4-(2-chlorophenyl)-4H-1,2,4-triazol-3-yl]-4,5-dihydrothieno[3,2-d][1]benzoxepin-8-yl}carbamate | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
5DXT
 
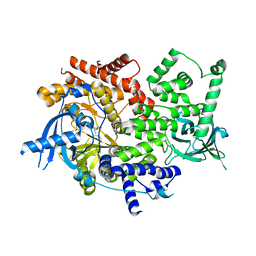 | | p110alpha with GDC-0326 | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, 1,2-ETHANEDIOL, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Heffron, T.P, Heald, R.A, Ndubaku, C, Wei, B.Q, Augustin, M, Do, S, Edgar, K, Eigenbrot, C, Friedman, L, Gancia, E, Jackson, P.S, Jones, G, Kolesnikov, A, Lee, L.B, Lesnick, J.D, Lewis, C, McLean, N, Mortle, M, Nonomiya, J, Pang, J, Price, S, Prior, W.W, Salphati, L, Sideris, S, Staben, S.T, Steinbacher, S, Tsui, V, Wallin, J, Sampath, D, Olivero, A. | | Deposit date: | 2015-09-23 | | Release date: | 2016-01-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Rational Design of Selective Benzoxazepin Inhibitors of the alpha-Isoform of Phosphoinositide 3-Kinase Culminating in the Identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326).
J.Med.Chem., 59, 2016
|
|
3U0O
 
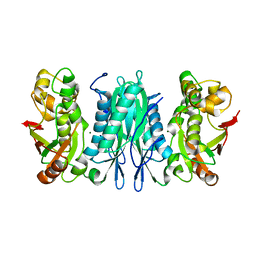 | | The crystal structure of selenophosphate synthetase from E. coli | | Descriptor: | MAGNESIUM ION, Selenide, water dikinase | | Authors: | Noinaj, N, Wattanasak, R, Wally, J, Stadtman, T, Buchanan, S.K. | | Deposit date: | 2011-09-28 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural insights into the catalytic mechanism of Escherichia coli selenophosphate synthetase.
J.Bacteriol., 194, 2012
|
|
4DQ2
 
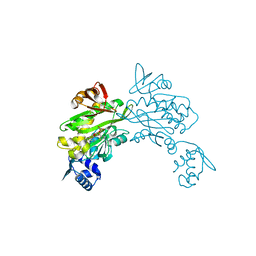 | | Structure of staphylococcus aureus biotin protein ligase in complex with biotinol-5'-amp | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, Biotin-[acetyl-CoA-carboxylase] ligase | | Authors: | Wilce, M, Yap, M, Pendini, N, Soares de Costa, T, Polyak, S, Tieu, W, Booker, G, Wallace, J. | | Deposit date: | 2012-02-14 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Selective inhibition of biotin protein ligase from Staphylococcus aureus.
J.Biol.Chem., 287, 2012
|
|
2MSR
 
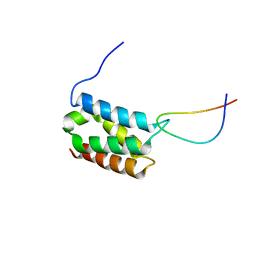 | | Solution structure of LEDGF/p75 IBD in complex with MLL1 peptide (140-160) | | Descriptor: | Histone-lysine N-methyltransferase 2A, PC4 and SFRS1-interacting protein | | Authors: | Cermakova, K, Tesina, P, Demeulemeester, J, El Ashkar, S, Mereau, H, Schwaller, J, Rezacova, P, Veverka, V, De Rijck, J. | | Deposit date: | 2014-08-05 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Validation and Structural Characterization of the LEDGF/p75-MLL Interface as a New Target for the Treatment of MLL-Dependent Leukemia.
Cancer Res., 74, 2014
|
|
1YQZ
 
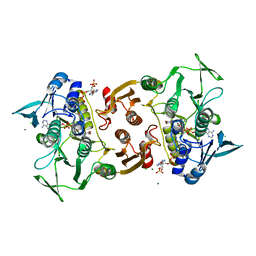 | | Structure of Coenzyme A-Disulfide Reductase from Staphylococcus aureus refined at 1.54 Angstrom resolution | | Descriptor: | CHLORIDE ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Mallett, T.C, Wallen, J.R, Sakai, H, Luba, J, Parsonage, D, Karplus, P.A, Tsukihara, T, Claiborne, A. | | Deposit date: | 2005-02-02 | | Release date: | 2006-05-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of coenzyme A-disulfide reductase from Staphylococcus aureus at 1.54 A resolution.
Biochemistry, 45, 2006
|
|
2V5P
 
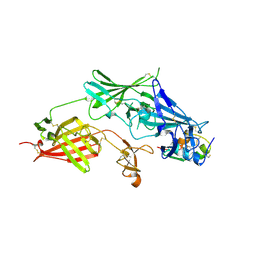 | | COMPLEX STRUCTURE OF HUMAN IGF2R DOMAINS 11-13 BOUND TO IGF-II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, INSULIN-LIKE GROWTH FACTOR II, ... | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
3BDO
 
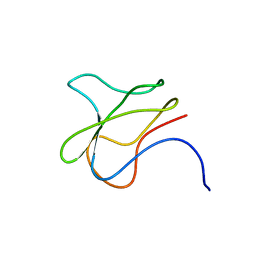 | | SOLUTION STRUCTURE OF APO-BIOTINYL DOMAIN FROM ACETYL COENZYME A CARBOXYLASE OF ESCHERICHIA COLI DETERMINED BY TRIPLE-RESONANCE NMR SPECTROSCOPY | | Descriptor: | PROTEIN (ACETYL-COA CARBOXYLASE) | | Authors: | Roberts, E.L, Shu, N, Howard, M.J, Broadhurst, R.W, Chapman-Smith, A, Wallace, J.C, Morris, T, Cronan, J.E, Perham, R.N. | | Deposit date: | 1999-03-08 | | Release date: | 1999-04-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of apo and holo biotinyl domains from acetyl coenzyme A carboxylase of Escherichia coli determined by triple-resonance nuclear magnetic resonance spectroscopy.
Biochemistry, 38, 1999
|
|
4PMW
 
 | |
2V5N
 
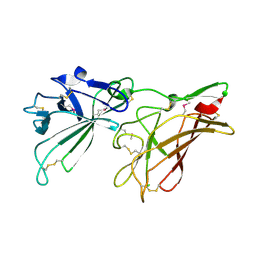 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
2V5O
 
 | | STRUCTURE OF HUMAN IGF2R DOMAINS 11-14 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATION-INDEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, CHLORIDE ION | | Authors: | Brown, J, Delaine, C, Zaccheo, O.J, Siebold, C, Gilbert, R.J, van Boxel, G, Denley, A, Wallace, J.C, Hassan, A.B, Forbes, B.E, Jones, E.Y. | | Deposit date: | 2007-07-06 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structure and Functional Analysis of the Igf-II/Igf2R Interaction
Embo J., 27, 2008
|
|
2JM2
 
 | | Structure of the N-terminal subdomain of insulin-like growth factor (IGF) binding protein-6 and its interactions with IGFs | | Descriptor: | Insulin-like growth factor-binding protein 6 | | Authors: | Chandrashekaran, I.R, Yao, S, Wang, C.C, Bansal, P.S, Alewood, P.F, Forbes, B.E, Wallace, J.C, Bach, L.A, Norton, R.S. | | Deposit date: | 2006-09-18 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | The N-Terminal Subdomain of Insulin-like Growth Factor (IGF) Binding Protein 6. Structure and Interaction with IGFs
Biochemistry, 46, 2007
|
|
