8JRD
 
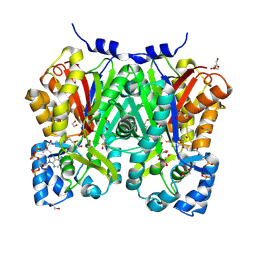 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin and coenzyme A | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Waki, T, Imaizumi, R, Nakata, S, Yanai, T, Takeshita, K, Sakai, N, Kataoka, K, Yamamoto, M, Nakayama, T, Yamashita, S. | | Deposit date: | 2023-06-16 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural insights into catalytic promiscuity of chalcone synthase from Glycine max (L.) Merr.: Coenzyme A-induced alteration of product specificity.
Biochem.Biophys.Res.Commun., 718, 2024
|
|
7BUR
 
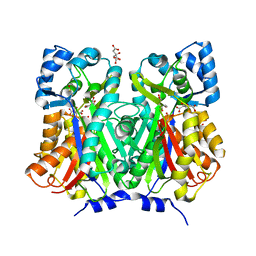 | | Chalcone synthase from Glycine max (L.) Merr (soybean) complexed with naringenin | | Descriptor: | CITRIC ACID, Chalcone synthase 1, NARINGENIN | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
7BUS
 
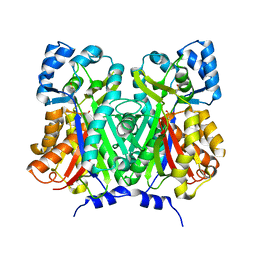 | | Chalcone synthase from Glycine max (L.) Merr (soybean) | | Descriptor: | Chalcone synthase | | Authors: | Imaizumi, R, Mameda, R, Takeshita, K, Waki, T, Kubo, H, Sakai, N, Nakata, S, Takahashi, S, Kataoka, K, Yamamoto, M, Yamashita, S, Nakayama, T. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of chalcone synthase, a key enzyme for isoflavonoid biosynthesis in soybean.
Proteins, 2020
|
|
6LMI
 
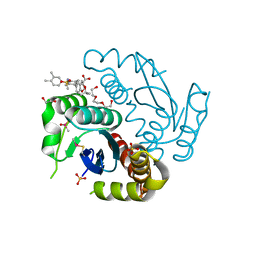 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1-benzopyran-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-chromen-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, Integrase catalytic, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-25 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
6LMQ
 
 | | Crystal structure of HIV-1 integrase catalytic core domain in complex with 2-(tert-butoxy)-2-[3-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-6-methanesulfonamido-2,3',4',5-tetramethyl-[1,1'-biphenyl]-4-yl]acetic acid | | Descriptor: | (2S)-2-[2-(3,4-dihydro-2H-1,4-benzoxazin-6-yl)-4-(3,4-dimethylphenyl)-3,6-dimethyl-5-(methylsulfonylamino)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, Integrase catalytic, SULFATE ION, ... | | Authors: | Sugiyama, S, Iwaki, T, Tamura, Y, Tomita, K, Matsuoka, E, Arita, S, Seki, T, Yoshinaga, T, Kawasuji, T. | | Deposit date: | 2019-12-26 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of novel integrase-LEDGF/p75 allosteric inhibitors based on a benzene scaffold.
Bioorg.Med.Chem., 28, 2020
|
|
3IYO
 
 | | Cryo-EM model of virion-sized HEV virion-sized capsid | | Descriptor: | Capsid protein | | Authors: | Xing, L, Mayazaki, N, Li, T.C, Simons, M.N, Wall, J.S, Moore, M, Wang, C.Y, Takeda, N, Wakita, T, Miyamura, T, Cheng, R.H. | | Deposit date: | 2010-03-19 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10.5 Å) | | Cite: | Structural basis for the RNA-dependent assembly pathway of hepatitis E virion-sized particles
J.Biol.Chem., 2010
|
|
7FCI
 
 | | human NTCP in complex with YN69083 Fab | | Descriptor: | Fab Heavy chain, Fab Light chain, Sodium/bile acid cotransporter | | Authors: | Park, J.H, Iwamoto, M, Yun, J.H, Uchikubo-Kamo, T, Son, D, Jin, Z, Yoshida, H, Ohki, M, Ishimoto, N, Mizutani, K, Oshima, M, Muramatsu, M, Wakita, T, Shirouzu, M, Liu, K, Uemura, T, Nomura, N, Iwata, S, Watashi, K, Tame, J.R.H, Nishizawa, T, Lee, W, Park, S.Y. | | Deposit date: | 2021-07-14 | | Release date: | 2022-05-25 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature, 606, 2022
|
|
1HNR
 
 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
6KS0
 
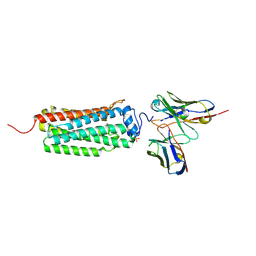 | | Crystal structure of the human adiponectin receptor 1 | | Descriptor: | Adiponectin receptor protein 1, PHOSPHATE ION, The heavy chain variable domain (Antibody), ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
6KS1
 
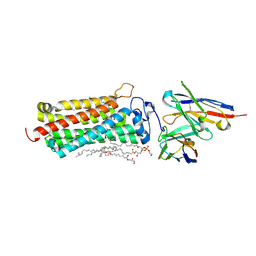 | | Crystal structure of the human adiponectin receptor 2 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-hydroxypropyl hexadecanoate, ... | | Authors: | Tanabe, H, Fujii, Y, Nakamura, Y, Hosaka, T, Okada-Iwabu, M, Iwabu, M, Kimura-Someya, T, Shirouzu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
6KRZ
 
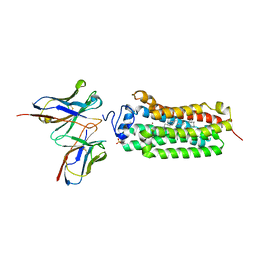 | | Crystal structure of the human adiponectin receptor 1 D208A mutant | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adiponectin receptor protein 1, OLEIC ACID, ... | | Authors: | Tanabe, H, Fujii, Y, Okada-Iwabu, M, Iwabu, M, Yamauchi, T, Kadowaki, T, Yokoyama, S. | | Deposit date: | 2019-08-22 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Human adiponectin receptor AdipoR1 assumes closed and open structures.
Commun Biol, 3, 2020
|
|
1HNS
 
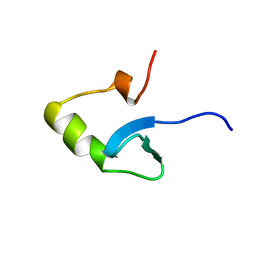 | | H-NS (DNA-BINDING DOMAIN) | | Descriptor: | H-NS | | Authors: | Shindo, H, Iwaki, T, Ieda, R, Kurumizaka, H, Ueguchi, C, Mizuno, T, Morikawa, S, Nakamura, H, Kuboniwa, H. | | Deposit date: | 1995-04-06 | | Release date: | 1995-07-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the DNA binding domain of a nucleoid-associated protein, H-NS, from Escherichia coli.
FEBS Lett., 360, 1995
|
|
1IT6
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN CALYCULIN A AND THE CATALYTIC SUBUNIT OF PROTEIN PHOSPHATASE 1 | | Descriptor: | CALYCULIN A, MANGANESE (II) ION, SERINE/THREONINE PROTEIN PHOSPHATASE 1 GAMMA (PP1-GAMMA) CATALYTIC SUBUNIT | | Authors: | Kita, A, Matsunaga, S, Takai, A, Kataiwa, H, Wakimoto, T, Fusetani, N, Isobe, M, Miki, K. | | Deposit date: | 2002-01-09 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the complex between calyculin A and the catalytic subunit of protein phosphatase 1.
Structure, 10, 2002
|
|
3ASQ
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with H-antigen | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASR
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-a | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASS
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3ASP
 
 | | Crystal structure of P domain from Norovirus Funabashi258 stain in the complex with A-antigen | | Descriptor: | Capsid protein, SODIUM ION, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
3AST
 
 | | Crystal structure of P domain Q389N mutant from Norovirus Funabashi258 stain in the complex with Lewis-b | | Descriptor: | Capsid protein, P-NITROPHENOL, SODIUM ION, ... | | Authors: | Kubota, T, Kumagai, A, Itoh, H, Furukawa, S, Narimatsu, H, Wakita, T, Ishii, K, Takeda, N, Someya, Y, Shirato, H. | | Deposit date: | 2010-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the recognition of Lewis antigens by genogroup I norovirus
J.Virol., 86, 2012
|
|
