3IPD
 
 | | Helical extension of the neuronal SNARE complex into the membrane, spacegroup I 21 21 21 | | Descriptor: | Synaptosomal-associated protein 25, Syntaxin-1A, Vesicle-associated membrane protein 2 | | Authors: | Stein, A, Weber, G, Wahl, M.C, Jahn, R. | | Deposit date: | 2009-08-17 | | Release date: | 2009-09-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Helical extension of the neuronal SNARE complex into the membrane
Nature, 460, 2009
|
|
6ZWX
 
 | | Crystal structure of E. coli RNA helicase HrpA | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZWW
 
 | | Crystal structure of E. coli RNA helicase HrpA in complex with RNA | | Descriptor: | ATP-dependent RNA helicase HrpA, CALCIUM ION, ssRNA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-07-29 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7AKP
 
 | | Crystal structure of E. coli RNA helicase HrpA-D305A | | Descriptor: | ATP-dependent RNA helicase HrpA | | Authors: | Grass, L.M, Wollenhaupt, J, Barthel, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-10-01 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Large-scale ratcheting in a bacterial DEAH/RHA-type RNA helicase that modulates antibiotics susceptibility.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7B3A
 
 | | Crystal structure of PamZ | | Descriptor: | ACETATE ION, ACETYL COENZYME *A, CHLORIDE ION, ... | | Authors: | Loll, B, Dang, T, Mainz, A, Suessmuth, R, Wahl, M.C. | | Deposit date: | 2020-11-30 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Molecular basis of antibiotic self-resistance in a bee larvae pathogen.
Nat Commun, 13, 2022
|
|
7BDK
 
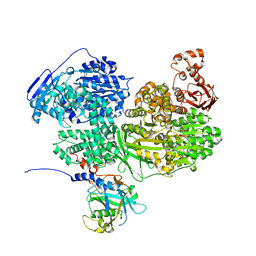 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDJ
 
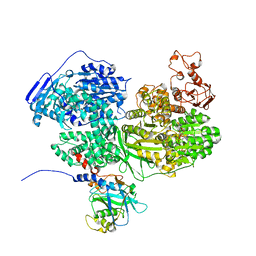 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ATPgammaS | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDL
 
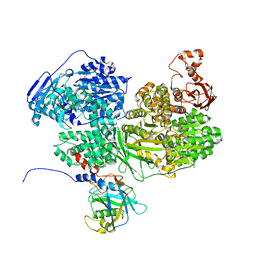 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ADP | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDI
 
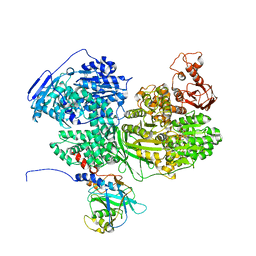 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
3LSA
 
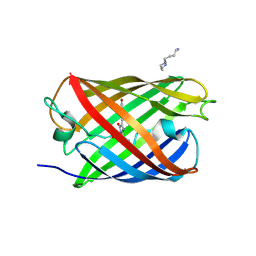 | | Padron0.9-OFF (non-fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
3LS3
 
 | | Padron0.9-ON (fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
4F92
 
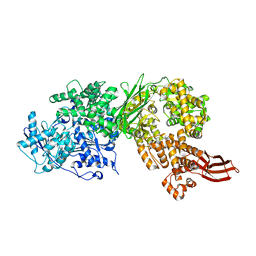 | | Brr2 Helicase Region S1087L | | Descriptor: | SULFANILAMIDE, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.662 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F91
 
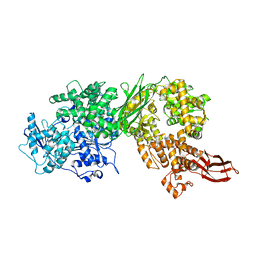 | | Brr2 Helicase Region | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F93
 
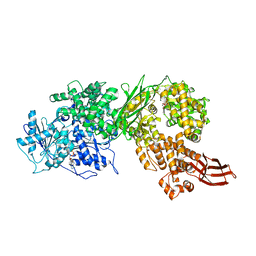 | | Brr2 Helicase Region S1087L, Mg-ATP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Santos, K.F, Jovin, S.M, Weber, G, Pena, V, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2012-05-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1WXG
 
 | | E.coli NAD Synthetase, DND | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXI
 
 | | E.coli NAD Synthetase, AMP.PP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXH
 
 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXE
 
 | | E.coli NAD Synthetase, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXF
 
 | | E.coli NAD Synthetase | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1ZAX
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form B | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZAW
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form A | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZAV
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P21 | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
2A54
 
 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A52
 
 | | fluorescent protein asFP595, S158V, on-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A50
 
 | | fluorescent protein asFP595, wt, off-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
