6QV4
 
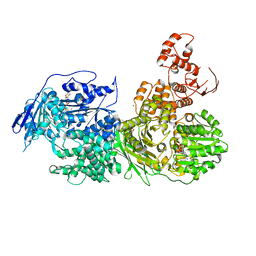 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
6QV3
 
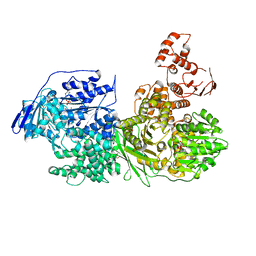 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
5T73
 
 | |
1WXG
 
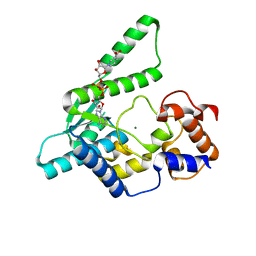 | | E.coli NAD Synthetase, DND | | Descriptor: | MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase, NICOTINIC ACID ADENINE DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXH
 
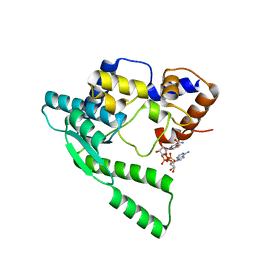 | | E.coli NAD Synthetase, NAD | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXE
 
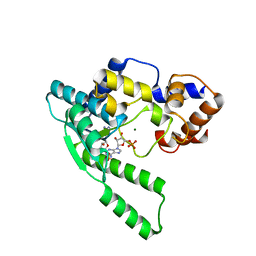 | | E.coli NAD Synthetase, AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
1WXF
 
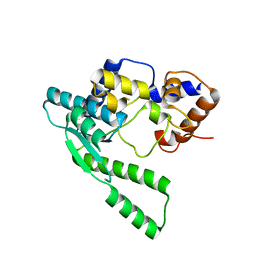 | | E.coli NAD Synthetase | | Descriptor: | NH(3)-dependent NAD(+) synthetase | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
6S8Q
 
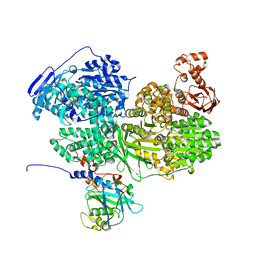 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, SULFATE ION, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Santos, K.F, Vester, K, Wahl, M.C. | | Deposit date: | 2019-07-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit.
J.Biol.Chem., 295, 2020
|
|
6S9I
 
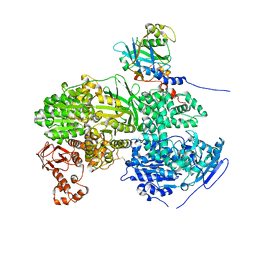 | |
6S8O
 
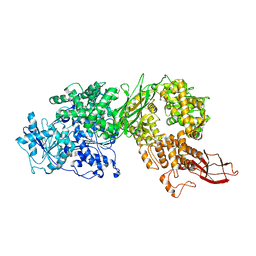 | | Human Brr2 Helicase Region M641C/A1582C | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Vester, K, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-07-10 | | Release date: | 2020-01-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.172 Å) | | Cite: | The inactive C-terminal cassette of the dual-cassette RNA helicase BRR2 both stimulates and inhibits the activity of the N-terminal helicase unit.
J.Biol.Chem., 295, 2020
|
|
1ZAX
 
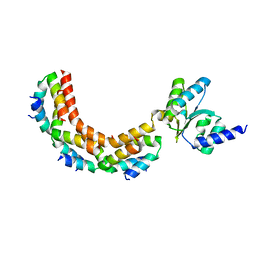 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form B | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
6TEO
 
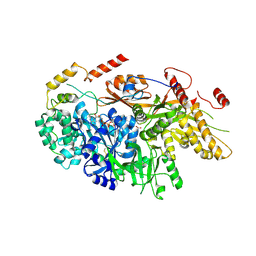 | | Crystal structure of a yeast Snu114-Prp8 complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-splicing factor 8, ... | | Authors: | Ganichkin, O, Jia, J, Loll, B, Absmeier, E, Wahl, M.C. | | Deposit date: | 2019-11-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Snu114-GTP-Prp8 module forms a relay station for efficient splicing in yeast.
Nucleic Acids Res., 48, 2020
|
|
6TQN
 
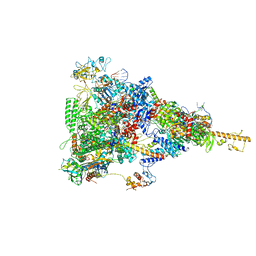 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
6TQO
 
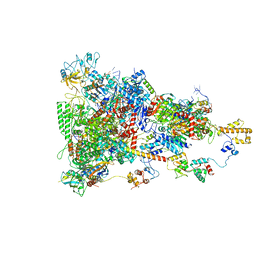 | | rrn anti-termination complex | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S4, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
1ZAV
 
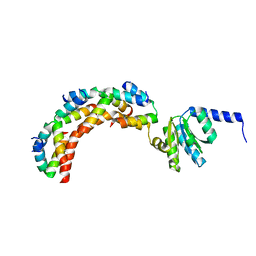 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P21 | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1ZAW
 
 | | Ribosomal Protein L10-L12(NTD) Complex, Space Group P212121, Form A | | Descriptor: | 50S ribosomal protein L10, 50S ribosomal protein L7/L12 | | Authors: | Diaconu, M, Kothe, U, Schluenzen, F, Fischer, N, Harms, J.M, Tonevitski, A.G, Stark, H, Rodnina, M.V, Wahl, M.C. | | Deposit date: | 2005-04-07 | | Release date: | 2005-07-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Function of the Ribosomal L7/12 Stalk in Factor Binding and GTPase Activation.
Cell(Cambridge,Mass.), 121, 2005
|
|
1WXI
 
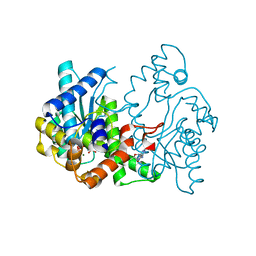 | | E.coli NAD Synthetase, AMP.PP | | Descriptor: | ADENOSINE MONOPHOSPHATE, DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jauch, R, Humm, A, Huber, R, Wahl, M.C. | | Deposit date: | 2005-01-23 | | Release date: | 2005-02-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Escherichia coli NAD Synthetase with Substrates and Products Reveal Mechanistic Rearrangements
J.Biol.Chem., 280, 2005
|
|
2A54
 
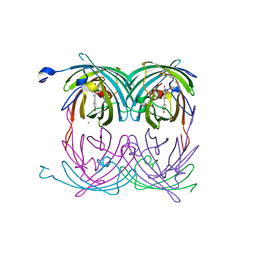 | | fluorescent protein asFP595, A143S, on-state, 1min irradiation | | Descriptor: | CHLORIDE ION, GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2A52
 
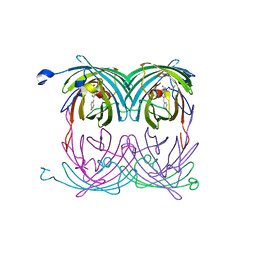 | | fluorescent protein asFP595, S158V, on-state | | Descriptor: | GFP-like non-fluorescent chromoprotein FP595 chain 1, GFP-like non-fluorescent chromoprotein FP595 chain 2 | | Authors: | Andresen, M, Wahl, M.C, Stiel, A.C, Graeter, F, Schaefer, L, Trowitzsch, S, Weber, G, Eggeling, C, Grubmueller, H, Hell, S.W, Jakobs, S. | | Deposit date: | 2005-06-30 | | Release date: | 2005-08-16 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the reversible photoswitch of a fluorescent protein
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
378D
 
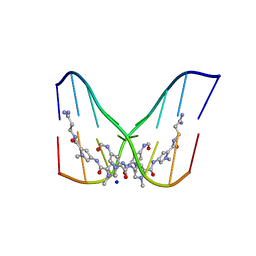 | | STRUCTURE OF THE SIDE-BY-SIDE BINDING OF DISTAMYCIN TO DNA | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*GP*TP*AP*TP*AP*TP*AP*C)-3'), SODIUM ION | | Authors: | Mitra, S.N, Wahl, M.C, Sundaralingam, M. | | Deposit date: | 1998-01-28 | | Release date: | 1999-03-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the side-by-side binding of distamycin to d(GTATATAC)2.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
2XHE
 
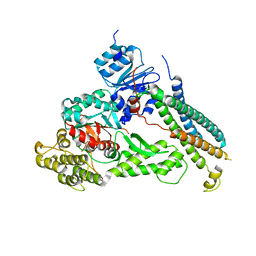 | | Crystal structure of the Unc18-syntaxin 1 complex from Monosiga brevicollis | | Descriptor: | SYNTAXIN1, UNC18 | | Authors: | Burkhardt, P, Stegmann, C.M, Wahl, M.C, Fasshauer, D. | | Deposit date: | 2010-06-14 | | Release date: | 2011-06-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Primordial Neurosecretory Apparatus Identified in the Choanoflagellate Monosiga Brevicollis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2XHC
 
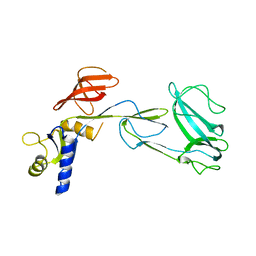 | |
2XHA
 
 | |
315D
 
 | |
3BC8
 
 | | Crystal structure of mouse selenocysteine synthase | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, O-phosphoseryl-tRNA(Sec) selenium transferase | | Authors: | Ganichkin, O.M, Wahl, M.C. | | Deposit date: | 2007-11-12 | | Release date: | 2007-12-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and catalytic mechanism of eukaryotic selenocysteine synthase.
J.Biol.Chem., 283, 2008
|
|
