6IB8
 
 | | Structure of a complex of SuhB and NusA AR2 domain | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, Inositol-1-monophosphatase, ... | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
6IB7
 
 | | Structure of wild type SuhB | | Descriptor: | GLYCEROL, Inositol-1-monophosphatase, MAGNESIUM ION | | Authors: | Huang, Y.H, Loll, B, Wahl, M.C. | | Deposit date: | 2018-11-29 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structural basis for the function of SuhB as a transcription factor in ribosomal RNA synthesis.
Nucleic Acids Res., 47, 2019
|
|
8Q3Q
 
 | |
8Q3O
 
 | |
8Q3N
 
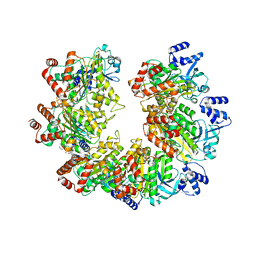 | |
8Q3P
 
 | |
6QWS
 
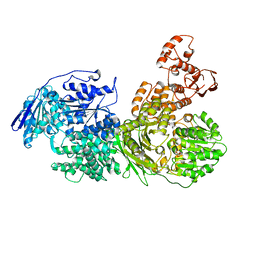 | |
6QV3
 
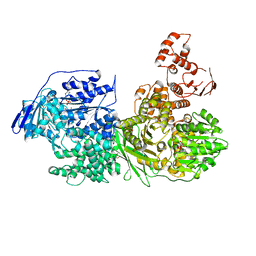 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
6QV4
 
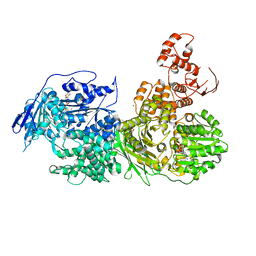 | | Crystal structure of the Ski2 RNA-helicase Brr2 from Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | ACETATE ION, MANGANESE (II) ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Absmeier, E, Santos, K.F, Wahl, M.C. | | Deposit date: | 2019-03-01 | | Release date: | 2020-01-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism Underlying Inhibition of Intrinsic ATPase Activity in a Ski2-like RNA Helicase.
Structure, 28, 2020
|
|
7OS1
 
 | | Cryo-EM structure of Brr2 in complex with Fbp21 | | Descriptor: | U5 small nuclear ribonucleoprotein 200 kDa helicase, WW domain-binding protein 4 | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
7OS2
 
 | | Cryo-EM structure of Brr2 in complex with Jab1/MPN and C9ORF78 | | Descriptor: | Pre-mRNA-processing-splicing factor 8, Telomere length and silencing protein 1 homolog, U5 small nuclear ribonucleoprotein 200 kDa helicase | | Authors: | Bergfort, A, Hilal, T, Weber, G, Wahl, M.C. | | Deposit date: | 2021-06-07 | | Release date: | 2022-02-23 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The intrinsically disordered TSSC4 protein acts as a helicase inhibitor, placeholder and multi-interaction coordinator during snRNP assembly and recycling.
Nucleic Acids Res., 50, 2022
|
|
6Z9T
 
 | | Transcription termination intermediate complex 5 | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9P
 
 | | Transcription termination intermediate complex 1 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9Q
 
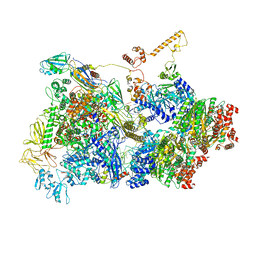 | | Transcription termination intermediate complex 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6YXQ
 
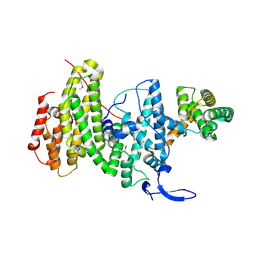 | | Crystal structure of a DNA repair complex ASCC3-ASCC2 | | Descriptor: | Activating signal cointegrator 1 complex subunit 2, Activating signal cointegrator 1 complex subunit 3 | | Authors: | Jia, J, Absmeier, E, Holton, N, Bohnsack, K.E, Pietrzyk-Brzezinska, A.J, Bohnsack, M.T, Wahl, M.C. | | Deposit date: | 2020-05-03 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The interaction of DNA repair factors ASCC2 and ASCC3 is affected by somatic cancer mutations.
Nat Commun, 11, 2020
|
|
6ZCA
 
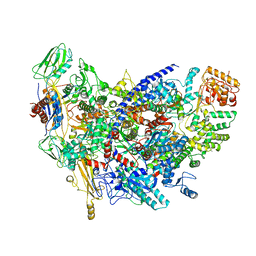 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (monomer) | | Descriptor: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-10 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
6Z9S
 
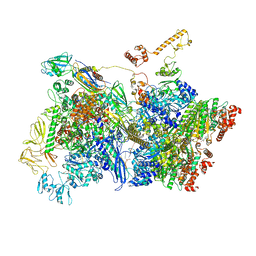 | | Transcription termination intermediate complex 4 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6Z9R
 
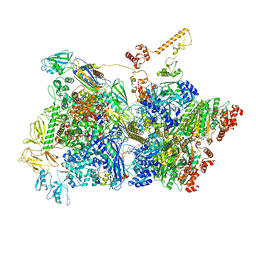 | | Transcription termination intermediate complex 3 | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Said, N, Hilal, T, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-04 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase rho.
Science, 371, 2021
|
|
6ZFB
 
 | | Structure of the B. subtilis RNA POLYMERASE in complex with HelD (dimer) | | Descriptor: | DNA helicase, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Pei, H.-P, Hilal, T, Huang, Y.-H, Said, N, Loll, B, Wahl, M.C. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The delta subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling.
Nat Commun, 11, 2020
|
|
7BDK
 
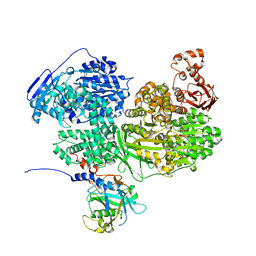 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDI
 
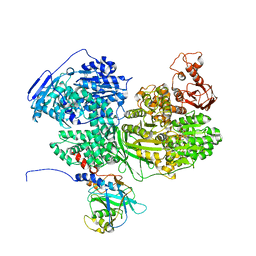 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Pre-mRNA-processing-splicing factor 8, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDJ
 
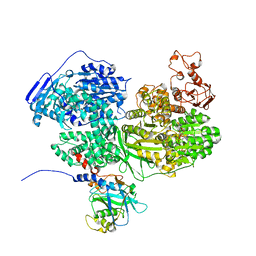 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ATPgammaS | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
7BDL
 
 | | Human Brr2 Helicase Region in complex with C-tail deleted Jab1 and mant-ADP | | Descriptor: | MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8, U5 small nuclear ribonucleoprotein 200 kDa helicase, ... | | Authors: | Vester, K, Santos, K.F, Absmeier, E, Wahl, M.C. | | Deposit date: | 2020-12-21 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Long-range allostery mediates cooperative adenine nucleotide binding by the Ski2-like RNA helicase Brr2.
J.Biol.Chem., 297, 2021
|
|
1TZU
 
 | | T. maritima NusB, P212121 | | Descriptor: | N utilization substance protein B homolog, SULFATE ION | | Authors: | Bonin, I, Robelek, R, Benecke, H, Urlaub, H, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-07-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the antitermination factor NusB from Thermotoga maritima and implications for RNA binding
Biochem.J., 383, 2004
|
|
6TW2
 
 | | Re-refined crystal structure of di-phosphorylated human CLK1 in complex with a novel substituted indole inhibitor | | Descriptor: | Dual specificity protein kinase CLK1, ethyl 3-[(E)-2-amino-1-cyanoethenyl]-6,7-dichloro-1-methyl-1H-indole-2-carboxylate | | Authors: | Loll, B, Haltenhof, T, Heyd, F, Wahl, M.C. | | Deposit date: | 2020-01-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Conserved Kinase-Based Body-Temperature Sensor Globally Controls Alternative Splicing and Gene Expression.
Mol.Cell, 78, 2020
|
|
