2RG8
 
 | |
2ROQ
 
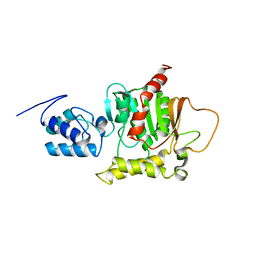 | | Solution Structure of the thiolation-thioesterase di-domain of enterobactin synthetase component F | | Descriptor: | Enterobactin synthetase component F | | Authors: | Frueh, D.P, Arthanari, H, Koglin, A, Vosburg, D.A, Bennett, A.E, Walsh, C.T, Wagner, G. | | Deposit date: | 2008-04-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase
Nature, 454, 2008
|
|
2RON
 
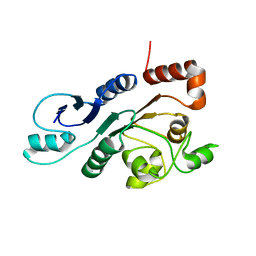 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
1KRI
 
 | |
7L0R
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0P
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, without AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7L0S
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, noncanonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1Y7Q
 
 | | Mammalian SCAN domain dimer is a domain-swapped homologue of the HIV capsid C-terminal domain | | Descriptor: | Zinc finger protein 174 | | Authors: | Ivanov, D, Stone, J.R, Maki, J.L, Collins, T, Wagner, G. | | Deposit date: | 2004-12-09 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Mammalian SCAN Domain Dimer Is a Domain-Swapped Homolog of the HIV Capsid C-Terminal Domain
Mol.Cell, 17, 2005
|
|
1XMW
 
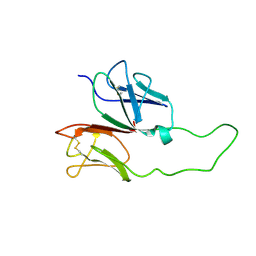 | | CD3 EPSILON AND DELTA ECTODOMAIN FRAGMENT COMPLEX IN SINGLE-CHAIN CONSTRUCT | | Descriptor: | Chimeric CD3 mouse Epsilon and sheep Delta Ectodomain Fragment Complex | | Authors: | Sun, Z.-Y.J, Kim, S.T, Kim, I.C, Fahmy, A, Reinherz, E.L, Wagner, G. | | Deposit date: | 2004-10-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the CD3epsilondelta ectodomain and comparison with CD3epsilongamma as a basis for modeling T cell receptor topology and signaling.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
7L0Q
 
 | | Structure of NTS-NTSR1-Gi complex in lipid nanodisc, canonical state, with AHD | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(T) subunit gamma-T1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Zhang, M, Gui, M, Wang, Z, Gorgulla, C, Yu, J.J, Wu, H, Sun, Z, Klenk, C, Merklinger, L, Morstein, L, Hagn, F, Pluckthun, A, Brown, A, Nasr, M.L, Wagner, G. | | Deposit date: | 2020-12-12 | | Release date: | 2021-01-06 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structure of an activated GPCR-G protein complex in lipid nanodiscs.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1ZGW
 
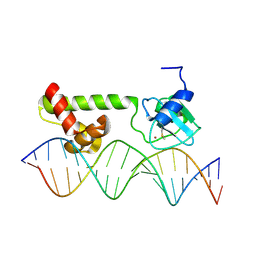 | | NMR structure of E. Coli Ada protein in complex with DNA | | Descriptor: | 5'-D(*GP*CP*AP*AP*AP*TP*TP*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*A)-3', 5'-D(*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*TP*AP*AP*TP*TP*TP*GP*C)-3', Ada polyprotein, ... | | Authors: | He, C, Hus, J.C, Sun, L.J, Zhou, P, Norman, D.P, Doetsch, V, Wei, H, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2005-04-22 | | Release date: | 2005-10-18 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | A Methylation-Dependent Electrostatic Switch Controls DNA Repair and Transcriptional Activation by E. coli Ada.
Mol.Cell, 20, 2005
|
|
1ZC1
 
 | | Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites | | Descriptor: | Ubiquitin fusion degradation protein 1 | | Authors: | Park, S, Isaacson, R, Kim, H.T, Silver, P.A, Wagner, G. | | Deposit date: | 2005-04-10 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Ufd1 Exhibits the AAA-ATPase Fold with Two Distinct Ubiquitin Interaction Sites
Structure, 13, 2005
|
|
1ADN
 
 | |
2ME3
 
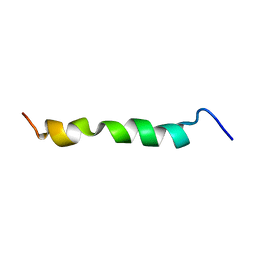 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
1BWM
 
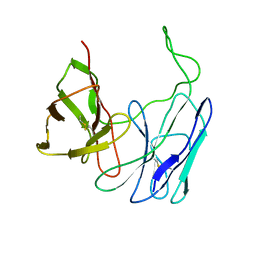 | | A SINGLE-CHAIN T CELL RECEPTOR | | Descriptor: | PROTEIN (ALPHA-BETA T CELL RECEPTOR (TCR) (D10)) | | Authors: | Hare, B.J, Wyss, D.F, Reinherz, E.L, Wagner, G. | | Deposit date: | 1998-09-23 | | Release date: | 1999-07-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure, specificity and CDR mobility of a class II restricted single-chain T-cell receptor.
Nat.Struct.Biol., 6, 1999
|
|
1CDB
 
 | |
2ME4
 
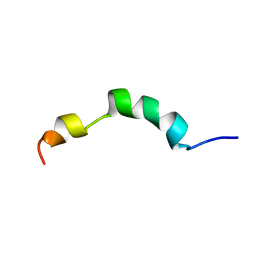 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME1
 
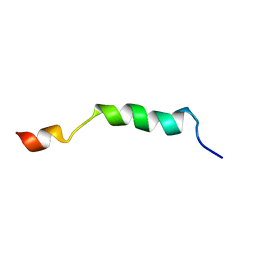 | | HIV-1 gp41 clade B double alanine mutant Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Gp41 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2ME2
 
 | | HIV-1 gp41 clade C Membrane Proximal External Region peptide in DPC micelle | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Sun, Z.J, Wagner, G, Reinherz, E.L, Kim, M, Song, L, Choi, J, Cheng, Y, Chowdhury, B, Bellot, G, Shih, W. | | Deposit date: | 2013-09-20 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Disruption of Helix-Capping Residues 671 and 674 Reveals a Role in HIV-1 Entry for a Specialized Hinge Segment of the Membrane Proximal External Region of gp41.
J.Mol.Biol., 426, 2014
|
|
2PAC
 
 | | SOLUTION STRUCTURE OF FE(II) CYTOCHROME C551 FROM PSEUDOMONAS AERUGINOSA AS DETERMINED BY TWO-DIMENSIONAL 1H NMR | | Descriptor: | CYTOCHROME C551, HEME C | | Authors: | Detlefsen, D.J, Thanabal, V, Pecoraro, V.L, Wagner, G. | | Deposit date: | 1993-05-05 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Fe(II) cytochrome c551 from Pseudomonas aeruginosa as determined by two-dimensional 1H NMR.
Biochemistry, 30, 1991
|
|
5GCN
 
 | | CATALYTIC DOMAIN OF TETRAHYMENA GCN5 HISTONE ACETYLTRANSFERASE IN COMPLEX WITH COENZYME A | | Descriptor: | COENZYME A, HISTONE ACETYLTRANSFERASE GCN5 | | Authors: | Lin, Y, Fletcher, C.M, Zhou, J, Allis, C.D, Wagner, G. | | Deposit date: | 1999-03-24 | | Release date: | 1999-07-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A
Nature, 400, 1999
|
|
1CSA
 
 | |
1I4V
 
 | |
1IIE
 
 | |
1J8C
 
 | | Solution Structure of the Ubiquitin-like Domain of hPLIC-2 | | Descriptor: | ubiquitin-like protein hPLIC-2 | | Authors: | Walters, K.J, Kleijnen, M.F, Goh, A.M, Wagner, G, Howley, P.M. | | Deposit date: | 2001-05-21 | | Release date: | 2002-03-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a.
Biochemistry, 41, 2002
|
|
