4M1J
 
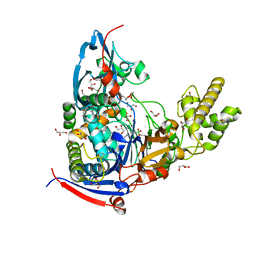 | | Crystal structure of Pseudomonas aeruginosa PvdQ in complex with a transition state analogue | | Descriptor: | Acyl-homoserine lactone acylase PvdQ subunit alpha, Acyl-homoserine lactone acylase PvdQ subunit beta, GLYCEROL, ... | | Authors: | Wu, R, Clevenger, K, Er, J, Fast, W.L, Liu, D. | | Deposit date: | 2013-08-02 | | Release date: | 2013-08-28 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Design of a Transition State Analogue with Picomolar Affinity for Pseudomonas aeruginosa PvdQ, a Siderophore Biosynthetic Enzyme.
Acs Chem.Biol., 8, 2013
|
|
4N05
 
 | |
4N04
 
 | |
3RAC
 
 | |
3RAG
 
 | |
4EWF
 
 | |
4M88
 
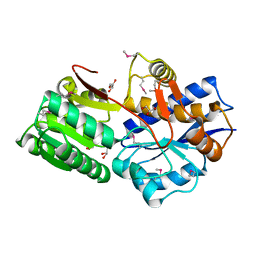 | |
4MAA
 
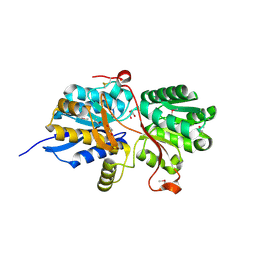 | |
5T4L
 
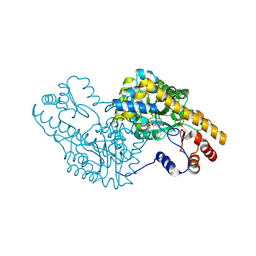 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4R)-4-amino-6-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}hexanoic acid, Aspartate aminotransferase | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4J
 
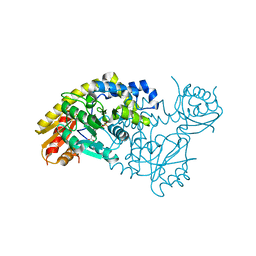 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | GAMMA-AMINO-BUTANOIC ACID, HTH-type transcriptional regulatory protein GabR, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.231 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5T4K
 
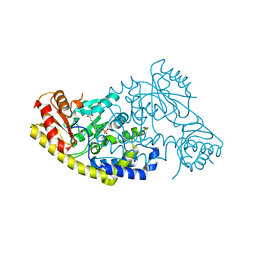 | | PLP and GABA Trigger GabR-Mediated Transcription Regulation in Bacillus subsidies via External Aldimine Formation | | Descriptor: | (4S)-5-fluoro-4-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]pentanoic acid, HTH-type transcriptional regulatory protein GabR | | Authors: | Wu, R, Sanishvili, R, Belitsky, B.R, Juncosa, J.I, Le, H.V, Lehrer, H.J.S, Farley, M, Silverman, R.B, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2016-08-29 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4N01
 
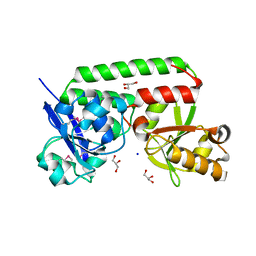 | |
5UBL
 
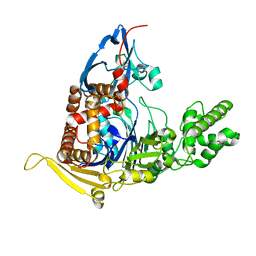 | | A circularly permuted version of PvdQ (cpPvdQ) | | Descriptor: | Acyl-homoserine lactone acylase PvdQ | | Authors: | Wu, R, Mascarenhas, R, Catlin, D, Clevenger, K, Fast, W, Liu, D. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-01 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Circular Permutation Reveals a Chromophore Precursor Binding Pocket of the Siderophore Tailoring Enzyme PvdQ
To Be Published
|
|
3NRT
 
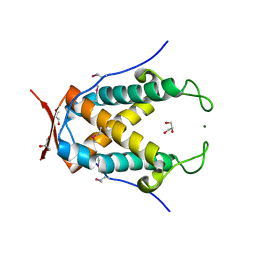 | | The crystal structure of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative ryanodine receptor | | Authors: | Wu, R, Zhang, R, James, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-30 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | he crystal strucutre of putative ryanodine receptor from Bacteroides thetaiotaomicron VPI-5482
To be Published
|
|
3ICY
 
 | |
1T5B
 
 | | Structural genomics, A protein from Salmonella typhimurium similar to E. coli acyl carrier protein phosphodiesterase | | Descriptor: | Acyl carrier protein phosphodiesterase, FLAVIN MONONUCLEOTIDE | | Authors: | Zhang, R, Wu, R, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-05-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 1.4 A crystal structure of a protein from Salmonella typhimurium similar to E. coli acyl carrier protein phosphodiesterase
To be Published
|
|
2IDL
 
 | | Crystal Structure of Conserved Protein of Unknown Function from Streptococcus pneumoniae | | Descriptor: | GLYCEROL, Hypothetical protein, SODIUM ION | | Authors: | Nocek, B, Wu, R, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-09-15 | | Release date: | 2006-10-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of conserved hypothetical protein from Streptococcus pneumoniae TIGR4
To be Published
|
|
1T06
 
 | |
3E7H
 
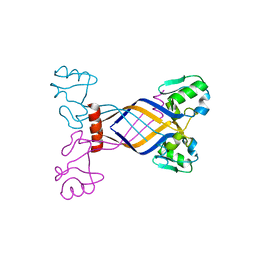 | |
4W9R
 
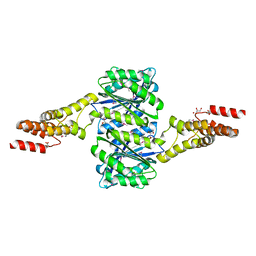 | | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271 | | Descriptor: | ACETATE ION, GLYCEROL, Uncharacterized protein | | Authors: | Chang, C, Wu, R, Clancy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.703 Å) | | Cite: | Crystal structure of uncharacterised protein Coch_1243 from Capnocytophaga ochracea DSM 7271
To Be Published
|
|
4S1P
 
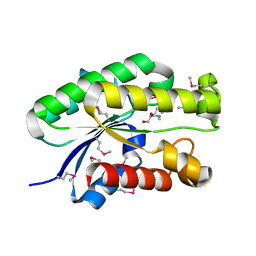 | | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens | | Descriptor: | UNKNOWN LIGAND, Uncharacterized protein | | Authors: | Osipiuk, J, Cuff, M.E, Wu, R, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-14 | | Release date: | 2015-01-28 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Shel_16390 protein, a putative SGNH hydrolase from Slackia heliotrinireducens
To be Published
|
|
6X1L
 
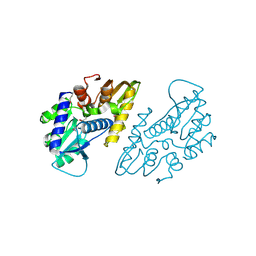 | | The crystal structure of a functional uncharacterized protein KP1_0663 from Klebsiella pneumoniae subsp. pneumoniae NTUH-K2044 | | Descriptor: | WbbZ protein | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-05-19 | | Release date: | 2020-06-03 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Systems Biology Approach to High-Risk CG23 Klebsiella pneumoniae.
Microbiol Resour Announc, 12, 2023
|
|
6X9Y
 
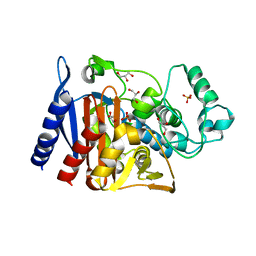 | | The crystal structure of a Beta-lactamase from Escherichia coli CFT073 | | Descriptor: | Beta-lactamase, GLYCEROL, S,R MESO-TARTARIC ACID, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of a Beta-lactamase from Escherichia coli CFT073
To Be Published
|
|
2GK3
 
 | | Cytoplasmic Protein STM3548 from Salmonella typhimurium | | Descriptor: | GLYCEROL, putative cytoplasmic protein | | Authors: | Petrova, T, Cuff, M.E, Wu, R.Y, Holzle, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-03-31 | | Release date: | 2006-05-02 | | Last modified: | 2011-08-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Novel hexamerization motif is discovered in a conserved cytoplasmic protein from Salmonella typhimurium.
J.Struct.Funct.Genom., 8, 2007
|
|
6WGQ
 
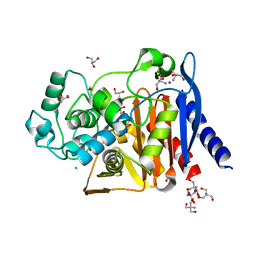 | | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Tan, K, Wu, R, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-06 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of a beta-lactamase from Shigella flexneri 2a str. 2457T
To Be Published
|
|
