5DP6
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 7 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-cyclopropylpropanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
8JMJ
 
 | | Structure of Helicobacter pylori Soj-DNA-Spo0J complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 2024
|
|
8JML
 
 | |
8JMK
 
 | | Structure of Helicobacter pylori Soj mutant, D41A bound to DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Wu, C.T, Chu, C.H, Sun, Y.J. | | Deposit date: | 2023-06-05 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into the molecular mechanism of ParABS system in chromosome partition by HpParA and HpParB.
Nucleic Acids Res., 2024
|
|
7XBI
 
 | | The crystal structure of human TrkA kinase bound to the inhibitor | | Descriptor: | 4-[[2-fluoranyl-5-(trifluoromethyl)phenyl]carbamoylamino]-~{N}-[3-(1-methylpyrazol-4-yl)-1~{H}-indazol-5-yl]-2-(trifluoromethyl)benzamide, CHLORIDE ION, High affinity nerve growth factor receptor | | Authors: | Wu, C.Y, Wang, G, Ouyang, L. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The crystal structure of human TrkA kinase bound to the inhibitor
To be published
|
|
8YTX
 
 | | Tubulin-RB3-TTL in complex with compound SI9 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[2,3-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Tubulin-RB3-TTL in complex with compound SI9
To Be Published
|
|
8ZB8
 
 | | Crystal structure of T2R-TTL-DPP21 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Detyrosinated tubulin alpha-1B chain, ... | | Authors: | Wu, C.Y, Chen, J.J. | | Deposit date: | 2024-04-26 | | Release date: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Crystal structure of T2R-TTL-DPP21 complex
To Be Published
|
|
8YUA
 
 | | Tubulin-RB3-TTL in complex with compound SI10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-chloranyl-~{N}-(4-methoxyphenyl)-~{N}-methyl-thieno[3,2-d]pyrimidin-4-amine, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The complex structure of tubulin-RB3-TTL in complex with compound SI10
To Be Published
|
|
8YU9
 
 | | Tubulin-RB3-TTL in complex with compound SI10 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-chloranylthieno[3,2-d]pyrimidin-4-yl)-7-methoxy-1,3-dihydroquinoxalin-2-one, CALCIUM ION, ... | | Authors: | Wu, C.Y, Wang, Y.X. | | Deposit date: | 2024-03-27 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The crystal structure of tubulin-RB3-TTL in complex with compound AB10
To Be Published
|
|
7B1R
 
 | |
7O2N
 
 | | Crystal structure of B. subtilis UGPase YngB | | Descriptor: | Probable UTP--glucose-1-phosphate uridylyltransferase YngB | | Authors: | Wu, C. | | Deposit date: | 2021-03-30 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Functional Characterisation of the Bacillus subtilis Uridylyltransferase YngB
Ph.D.Thesis, 2021
|
|
2VOY
 
 | | CryoEM model of CopA, the copper transporting ATPase from Archaeoglobus fulgidus | | Descriptor: | CATION-TRANSPORTING ATPASE, P-TYPE, POTENTIAL COPPER-TRANSPORTING ATPASE, ... | | Authors: | Wu, C.-C, Rice, W.J, Stokes, D.L. | | Deposit date: | 2008-02-25 | | Release date: | 2009-05-26 | | Last modified: | 2022-05-04 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of a Copper Pump Suggests a Regulatory Role for its Metal-Binding Domain.
Structure, 16, 2008
|
|
1HRK
 
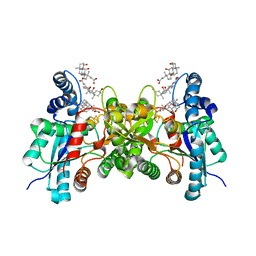 | | CRYSTAL STRUCTURE OF HUMAN FERROCHELATASE | | Descriptor: | CHOLIC ACID, FE2/S2 (INORGANIC) CLUSTER, FERROCHELATASE | | Authors: | Wu, C.K, Dailey, H.A, Rose, J.P, Burden, A, Sellers, V.M, Wang, B.-C. | | Deposit date: | 2000-12-21 | | Release date: | 2001-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis.
Nat.Struct.Biol., 8, 2001
|
|
1YGZ
 
 | | Crystal Structure of Inorganic Pyrophosphatase from Helicobacter pylori | | Descriptor: | Inorganic pyrophosphatase | | Authors: | Wu, C.A, Lokanath, N.K, Kim, D.Y, Park, H.J, Hwang, H.Y, Kim, S.T, Suh, S.W, Kim, K.K. | | Deposit date: | 2005-01-06 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of inorganic pyrophosphatase from Helicobacter pylori.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
4ANI
 
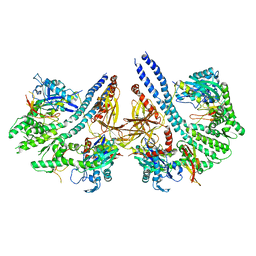 | | Structural basis for the intermolecular communication between DnaK and GrpE in the DnaK chaperone system from Geobacillus kaustophilus HTA426 | | Descriptor: | CHAPERONE PROTEIN DNAK, PROTEIN GRPE | | Authors: | Wu, C.-C, Naveen, V, Chien, C.-H, Chang, Y.-W, Hsiao, C.-D. | | Deposit date: | 2012-03-19 | | Release date: | 2012-05-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.094 Å) | | Cite: | Crystal Structure of Dnak Protein Complexed with Nucleotide Exchange Factor Grpe in Dnak Chaperone System: Insight Into Intermolecular Communication.
J.Biol.Chem., 287, 2012
|
|
5DP4
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 3 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2S)-2-methyl-3-phenylpropanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP9
 
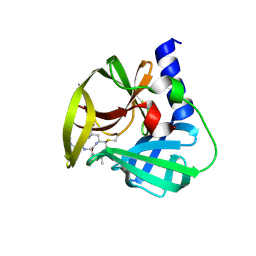 | | Crystal Structure of EV71 3C Proteinase in complex with compound 9 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(cyclobutylmethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP8
 
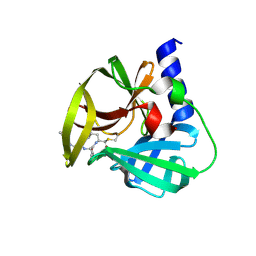 | | Crystal Structure of EV71 3C Proteinase in complex with compound 8 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-{[(2-cyclopropylethyl)amino](oxo)acetyl}-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DPA
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 6 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-[(N-acetyl-L-phenylalanyl)amino]-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP5
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 4 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[(2R,5S)-5-amino-2-(4-fluorobenzyl)-6-methyl-4-oxoheptanoyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2016-04-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP7
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 5 | | Descriptor: | 3C proteinase, ethyl (2Z,4S)-4-{[N-(3-methylbutanoyl)-L-phenylalanyl]amino}-5-[(3S)-2-oxopyrrolidin-3-yl]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
5DP3
 
 | | Crystal Structure of EV71 3C Proteinase in complex with compound 2 | | Descriptor: | 3C proteinase, ethyl (4S)-5-[(3S)-2-oxopyrrolidin-3-yl]-4-[(3-phenylpropanoyl)amino]pent-2-enoate | | Authors: | Wu, C, Zhang, L, Li, P, Cai, Q, Peng, X, Li, N, Cai, Y, Li, J, Lin, T. | | Deposit date: | 2015-09-12 | | Release date: | 2016-03-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Fragment-wise design of inhibitors to 3C proteinase from enterovirus 71
Biochim.Biophys.Acta, 1860, 2016
|
|
8XA9
 
 | | Human MGME1 in complex with 5'-overhang DNA | | Descriptor: | CALCIUM ION, DNA (11-MER), DNA (18-MER), ... | | Authors: | Wu, C.C, Mao, E.Y.C. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis of how MGME1 processes DNA 5' ends to maintain mitochondrial genome integrity.
Nucleic Acids Res., 52, 2024
|
|
3J08
 
 | |
3J09
 
 | |
