5WDE
 
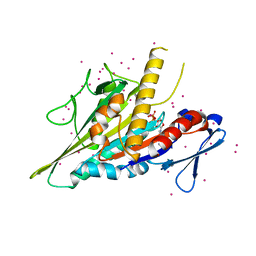 | | Crystal structure of the KIFC3 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIFC3, MAGNESIUM ION, ... | | Authors: | Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-07-05 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis of small molecule ATPase inhibition of a human mitotic kinesin motor protein.
Sci Rep, 7, 2017
|
|
8AZG
 
 | |
1PPE
 
 | |
2C5D
 
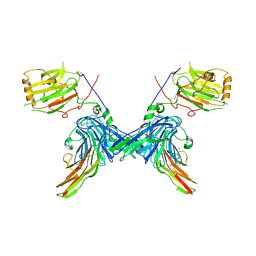 | | Structure of a minimal Gas6-Axl complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN 6 PRECURSOR, ... | | Authors: | Sasaki, T, Knyazev, P.G, Clout, N.J, Cheburkin, Y, Goehring, W, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Gas6-Axl Signalling.
Embo J., 25, 2006
|
|
7N46
 
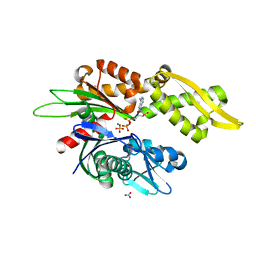 | |
2M0Y
 
 | |
1G1L
 
 | | THE STRUCTURAL BASIS OF THE CATALYTIC MECHANISM AND REGULATION OF GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE (RMLA). TDP-GLUCOSE COMPLEX. | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, CITRIC ACID, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Blankenfeldt, W, Asuncion, M, Lam, J.S, Naimsmith, J.H. | | Deposit date: | 2000-10-12 | | Release date: | 2000-12-27 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA).
EMBO J., 19, 2000
|
|
7NPT
 
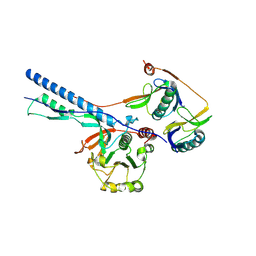 | | Cytosolic bridge of an intact ESX-5 inner membrane complex | | Descriptor: | ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-26 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
7NPS
 
 | | Structure of the periplasmic assembly from the ESX-5 inner membrane complex, C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, Mycosin-5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-05-26 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
7NP7
 
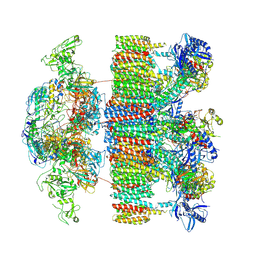 | | Structure of an intact ESX-5 inner membrane complex, Composite C1 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
4NV5
 
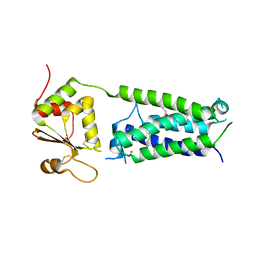 | | C50A mutant of Synechococcus VKOR, C2 crystal form (dehydrated) | | Descriptor: | UBIQUINONE-10, VKORC1/thioredoxin domain protein | | Authors: | Liu, S, Cheng, W, Fowle Grider, R, Shen, G, Li, W. | | Deposit date: | 2013-12-04 | | Release date: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer.
Nat Commun, 5, 2014
|
|
8BCI
 
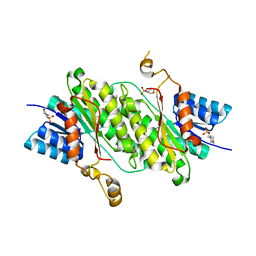 | |
7NPU
 
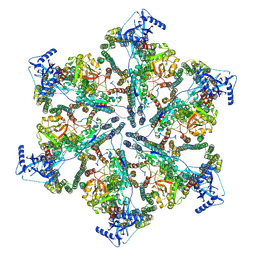 | | MycP5-free ESX-5 inner membrane complex, state I | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (4.48 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
8BCJ
 
 | |
7NPR
 
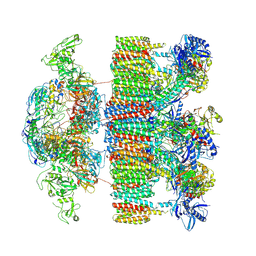 | | Structure of an intact ESX-5 inner membrane complex, Composite C3 model | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5, ... | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
7NPV
 
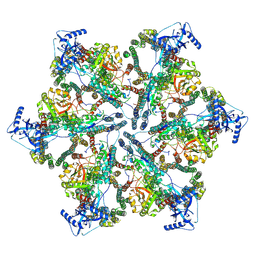 | | MycP5-free ESX-5 inner membrane complex, State II | | Descriptor: | ESX-5 secretion system ATPase EccB5, ESX-5 secretion system protein EccC5, ESX-5 secretion system protein EccD5 | | Authors: | Fahrenkamp, D, Bunduc, C.M, Wald, J, Ummels, R, Bitter, W, Houben, E.N.G, Marlovits, T.C. | | Deposit date: | 2021-02-28 | | Release date: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Structure and dynamics of a mycobacterial type VII secretion system.
Nature, 593, 2021
|
|
3IUF
 
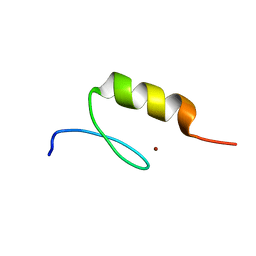 | | Crystal structure of the C2H2-type zinc finger domain of human ubi-d4 | | Descriptor: | ZINC ION, Zinc finger protein ubi-d4 | | Authors: | Tempel, W, Xu, C, Bian, C, Adams-Cioaba, M, Eryilmaz, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-11-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Cys2His2-type zinc finger domain of human DPF2.
Biochem.Biophys.Res.Commun., 413, 2011
|
|
4NTJ
 
 | | Structure of the human P2Y12 receptor in complex with an antithrombotic drug | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CHOLESTEROL, P2Y purinoceptor 12,Soluble cytochrome b562,P2Y purinoceptor 12, ... | | Authors: | Zhang, K, Zhang, J, Gao, Z.-G, Zhang, D, Zhu, L, Han, G.W, Moss, S.M, Paoletta, S, Kiselev, E, Lu, W, Fenalti, G, Zhang, W, Muller, C.E, Yang, H, Jiang, H, Cherezov, V, Katritch, V, Jacobson, K.A, Stevens, R.C, Wu, B, Zhao, Q, GPCR Network (GPCR) | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structure of the human P2Y12 receptor in complex with an antithrombotic drug
Nature, 509, 2014
|
|
1OGT
 
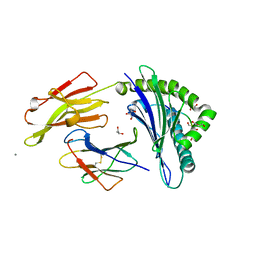 | | CRYSTAL STRUCTURE OF HLA-B*2705 COMPLEXED WITH THE VASOACTIVE INTESTINAL PEPTIDE TYPE 1 RECEPTOR (VIPR) PEPTIDE (RESIDUES 400-408) | | Descriptor: | BETA-2-MICROGLOBULIN, GLYCEROL, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ... | | Authors: | Hulsmeyer, M, Fiorillo, M.T, Bettosini, F, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, B. | | Deposit date: | 2003-05-13 | | Release date: | 2004-01-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Dual, HLA-B27 subtype-dependent conformation of a self-peptide.
J. Exp. Med., 199, 2004
|
|
8B4A
 
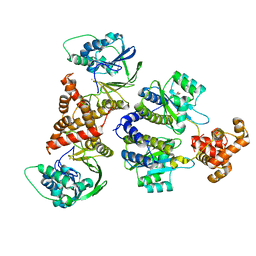 | | Nativ complex of PqsE and RhlR with autoinducer C4-HSL | | Descriptor: | 2-aminobenzoylacetyl-CoA thioesterase, FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, ... | | Authors: | Borgert, S.R, Blankenfeldt, W. | | Deposit date: | 2022-09-20 | | Release date: | 2022-12-14 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Moonlighting chaperone activity of the enzyme PqsE contributes to RhlR-controlled virulence of Pseudomonas aeruginosa.
Nat Commun, 13, 2022
|
|
1JAO
 
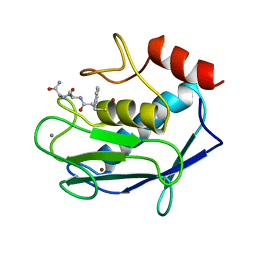 | | COMPLEX OF 3-MERCAPTO-2-BENZYLPROPANOYL-ALA-GLY-NH2 WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), N-[(2S)-2-benzyl-3-sulfanylpropanoyl]-L-alanylglycinamide, ... | | Authors: | Grams, F, Reinemer, P, Powers, J.C, Kleine, T, Piper, M, Tschesche, H, Huber, R, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structures of human neutrophil collagenase complexed with peptide hydroxamate and peptide thiol inhibitors. Implications for substrate binding and rational drug design.
Eur.J.Biochem., 228, 1995
|
|
1JAP
 
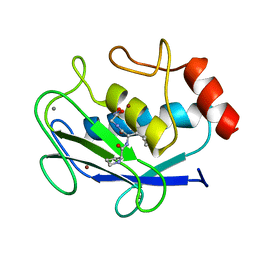 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (MET80 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (MET80 FORM), PRO-LEU-GLY-HYDROXYLAMINE, ... | | Authors: | Bode, W, Reinemer, P, Huber, R, Kleine, T, Schnierer, S, Tschesche, H. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | The X-ray crystal structure of the catalytic domain of human neutrophil collagenase inhibited by a substrate analogue reveals the essentials for catalysis and specificity.
EMBO J., 13, 1994
|
|
1JAN
 
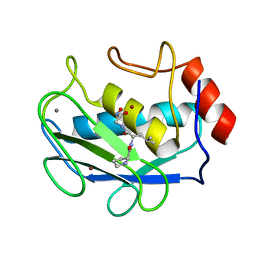 | | COMPLEX OF PRO-LEU-GLY-HYDROXYLAMINE WITH THE CATALYTIC DOMAIN OF MATRIX METALLO PROTEINASE-8 (PHE79 FORM) | | Descriptor: | CALCIUM ION, MATRIX METALLO PROTEINASE-8 (PHE79 FORM), PRO-LEU-GLY-HYDROXYLAMINE INHIBITOR, ... | | Authors: | Reinemer, P, Grams, F, Huber, R, Kleine, T, Schnierer, S, Pieper, M, Tschesche, H, Bode, W. | | Deposit date: | 1996-03-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural implications for the role of the N terminus in the 'superactivation' of collagenases. A crystallographic study.
FEBS Lett., 338, 1994
|
|
8BGN
 
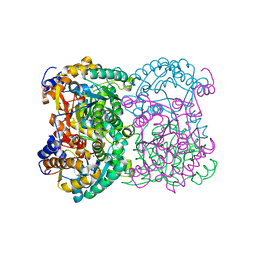 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Diacetylchitobiose deacetylase, ... | | Authors: | Rypniewski, W, Bejger, M, Biniek-Antosiak, K. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
8BGP
 
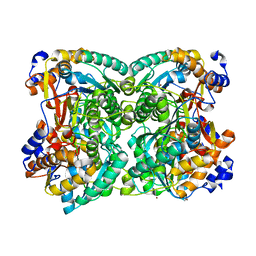 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus anomalous data | | Descriptor: | Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Biniek-Antosiak, K, Bejger, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
