3O3M
 
 | | (R)-2-Hydroxyisocaproyl-CoA Dehydratase | | Descriptor: | HYDROSULFURIC ACID, IRON/SULFUR CLUSTER, SULFATE ION, ... | | Authors: | Knauer, S.H, Buckel, W, Dobbek, H. | | Deposit date: | 2010-07-25 | | Release date: | 2011-03-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis for Reductive Radical Formation and Electron Recycling in (R)-2-Hydroxyisocaproyl-CoA Dehydratase.
J.Am.Chem.Soc., 133, 2011
|
|
4NWQ
 
 | | Computationally Designed Two-Component Self-Assembling Tetrahedral Cage, T33-21, Crystallized in Space Group F4132 | | Descriptor: | Putative uncharacterized protein PH0671, SULFATE ION, Uncharacterized protein | | Authors: | McNamara, D.E, King, N.P, Bale, J.B, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2013-12-06 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Accurate design of co-assembling multi-component protein nanomaterials.
Nature, 510, 2014
|
|
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1MZP
 
 | | Structure of the L1 protuberance in the ribosome | | Descriptor: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | Authors: | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | Deposit date: | 2002-10-09 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
3ZI1
 
 | | Crystal structure of human glyoxalase domain-containing protein 4 (GLOD4) | | Descriptor: | 1,2-ETHANEDIOL, GLYOXALASE DOMAIN-CONTAINING PROTEIN 4 | | Authors: | Oberholzer, A, Kiyani, W, Shrestha, L, Vollmar, M, Krojer, T, Froese, D.S, Williams, E, von Delft, F, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W.W. | | Deposit date: | 2012-12-30 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Human Glyoxalase Domain- Containing Protein 4 (Glod4)
To be Published
|
|
4NEK
 
 | | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, Enoyl-CoA hydratase/carnithine racemase | | Authors: | Tkaczuk, K.L, Cooper, D.R, Geffken, K, Chapman, H.C, Stead, M, Hillerich, B, Ahmed, M, Bonanno, J.B, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-29 | | Release date: | 2013-11-27 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Putative enoyl-CoA hydratase/carnithine racemase from Magnetospirillum magneticum AMB-1
To be Published
|
|
4NX2
 
 | | Crystal structure of DCYRS complexed with DCY | | Descriptor: | 3,5-dichloro-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Wang, J, Gong, W, Li, J, Gao, F, Li, H. | | Deposit date: | 2013-12-08 | | Release date: | 2014-09-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Significant expansion of fluorescent protein sensing ability through the genetic incorporation of superior photo-induced electron-transfer quenchers.
J.Am.Chem.Soc., 136, 2014
|
|
5Y29
 
 | | Crystal structure of Ostrinia furnacalis Group II chitinase catalytic domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, insect group II chitinase | | Authors: | Chen, W, Qu, M.B, Zhou, Y, Yang, Q. | | Deposit date: | 2017-07-24 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of group II chitinase (ChtII) catalysis completes the puzzle of chitin hydrolysis in insects
J. Biol. Chem., 293, 2018
|
|
3O1K
 
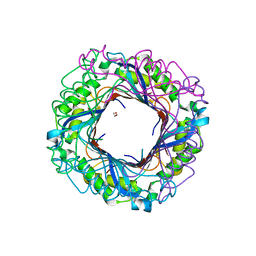 | | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961 | | Descriptor: | 1,2-ETHANEDIOL, Dihydroneopterin aldolase FolB, putative | | Authors: | Nocek, B, Zhou, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-21 | | Release date: | 2010-09-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of putative dihydroneopterin aldolase (FolB) from Vibrio cholerae O1 biovar El Tor str. N16961
To be Published
|
|
3ZQ0
 
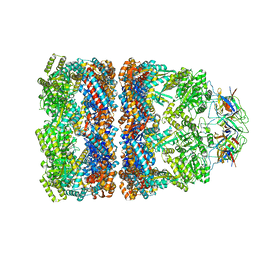 | | Visualizing GroEL-ES in the Act of Encapsulating a Non-Native Substrate Protein | | Descriptor: | 10 KDA CHAPERONIN, 60 KDA CHAPERONIN, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Chen, D.-H, Madan, D, Weaver, J, Lin, Z, Schroder, G.F, Chiu, W, Rye, H.S. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9.2 Å) | | Cite: | Visualizing Groel/Es in the Act of Encapsulating a Folding Protein
Cell(Cambridge,Mass.), 153, 2013
|
|
5UEG
 
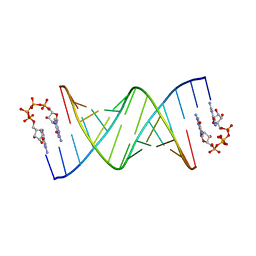 | | RNA primer-template complex with guanosine dinucleotide G(5')pppp(5')G ligand | | Descriptor: | RNA (5'-R(*(LCC)P*(LCC)P*(LCC)P*GP*AP*CP*UP*UP*AP*AP*GP*UP*CP*G)-3'), [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [[[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl] hydrogen phosphate | | Authors: | Zhang, W, Tam, C.P, Szostak, J.W. | | Deposit date: | 2017-01-02 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insight into the mechanism of nonenzymatic RNA primer extension from the structure of an RNA-GpppG complex.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4NHD
 
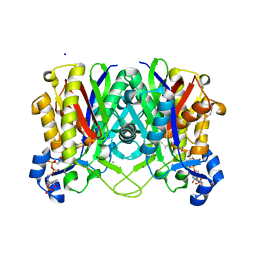 | | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] synthase 3 protein 1, CALCIUM ION, COENZYME A, ... | | Authors: | Hou, J, Zheng, H, Langner, K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-04 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of beta-ketoacyl-ACP synthase III (FabH) from Vibrio Cholerae in complex with Coenzyme A
To be Published
|
|
1NHJ
 
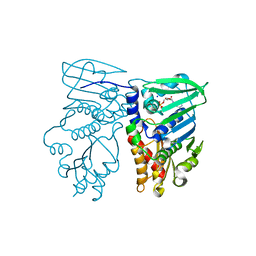 | | Crystal structure of N-terminal 40KD MutL/A100P mutant protein complex with ADPnP and one sodium | | Descriptor: | DNA mismatch repair protein mutL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
5TRU
 
 | | Structure of the first-in-class checkpoint inhibitor Ipilimumab bound to human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, Ipilimumab Fab heavy chain, Ipilimumab Fab light chain | | Authors: | Ramagopal, U.A, Liu, W, Garrett-Thomson, S.C, Yan, Q, Srinivasan, M, Wong, S.C, Bell, A, Mankikar, S, Rangan, V.S, Deshpande, S, Bonanno, J.B, Korman, A.J, Almo, S.C. | | Deposit date: | 2016-10-27 | | Release date: | 2017-05-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for cancer immunotherapy by the first-in-class checkpoint inhibitor ipilimumab.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1NA4
 
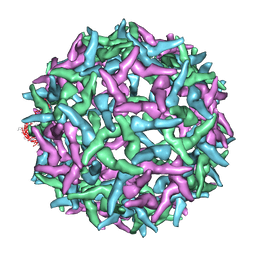 | | The structure of immature Yellow Fever virus particle | | Descriptor: | major envelope protein E | | Authors: | Zhang, Y, Corver, J, Chipman, P.R, Lenches, E, Zhang, W, Pletnev, S.V, Sedlak, D, Baker, T.S, Strauss, J.H, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2002-11-26 | | Release date: | 2003-12-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY | | Cite: | Structures of immature flavivirus particles
EMBO J., 22, 2003
|
|
5UEF
 
 | |
3ORR
 
 | | Crystal Structure of N5-Carboxyaminoimidazole synthetase from Staphylococcus aureus | | Descriptor: | N5-carboxyaminoimidazole ribonucleotide synthetase | | Authors: | Brugarolas, P, Duguid, E.M, Zhang, W, Poor, C.B, He, C. | | Deposit date: | 2010-09-07 | | Release date: | 2011-07-20 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical characterization of N5-carboxyaminoimidazole ribonucleotide synthetase and N5-carboxyaminoimidazole ribonucleotide mutase from Staphylococcus aureus.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
5YET
 
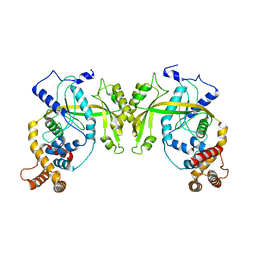 | | Structure of R354_WT | | Descriptor: | Uncharacterized protein R354 | | Authors: | Dou, C, Yu, M.J, Gu, Y.J, Cheng, W. | | Deposit date: | 2017-09-19 | | Release date: | 2018-06-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structural and Mechanistic Analyses Reveal a Unique Cas4-like Protein in the Mimivirus Virophage Resistance Element System.
Iscience, 3, 2018
|
|
5YF6
 
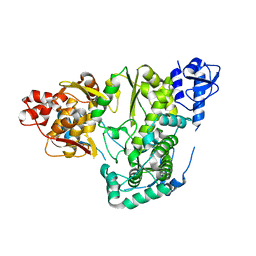 | |
5U2K
 
 | | Crystal structure of Galactoside O-acetyltransferase complex with CoA (H3 space group) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Czub, M.P, Porebski, P.J, Knapik, A.A, Niedzialkowska, E, Siuda, M.K, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-30 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of Galactoside O-acetyltransferase complex
with CoA (H3 space group)
to be published
|
|
5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5YP1
 
 | | Crystal structure of dipeptidyl peptidase IV (DPP IV) from Pseudoxanthomonas mexicana WO24 | | Descriptor: | Dipeptidyl aminopeptidase 4, GLYCEROL | | Authors: | Roppongi, S, Suzuki, Y, Tateoka, C, Fuimoto, M, Morisawa, S, Iizuka, I, Nakamura, A, Honma, N, Shida, Y, Ogasawara, W, Tanaka, N, Sakamoto, Y, Nonaka, T. | | Deposit date: | 2017-11-01 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Crystal structures of a bacterial dipeptidyl peptidase IV reveal a novel substrate recognition mechanism distinct from that of mammalian orthologues.
Sci Rep, 8, 2018
|
|
4ALB
 
 | | Structure of Phenolic Acid Decarboxylase from Bacillus subtilis: Tyr19Ala mutant in complex with coumaric acid | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHENOLIC ACID DECARBOXYLASE PADC | | Authors: | Frank, A, Eborall, W, Hyde, R, Hart, S, Turkenburg, J.P, Grogan, G. | | Deposit date: | 2012-03-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Mutational Analysis of Phenolic Acid Decarboxylase from Bacillus Subtilis (Bspad), which Converts Bio-Derived Phenolic Acids to Styrene Derivatives
Catal.Sci.Technol., 2, 2012
|
|
1NHH
 
 | | Crystal structure of N-terminal 40KD MutL protein (LN40) complex with ADPnP and one Rubidium | | Descriptor: | 1,2-ETHANEDIOL, DNA mismatch repair protein mutL, MAGNESIUM ION, ... | | Authors: | Hu, X, Machius, M, Yang, W. | | Deposit date: | 2002-12-19 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Monovalent cation dependence and preference of GHKL ATPases and kinases
FEBS Lett., 544, 2003
|
|
5YIM
 
 | | Structure of a Legionella effector | | Descriptor: | SdeA | | Authors: | Feng, Y, Dong, Y, Wang, W. | | Deposit date: | 2017-10-05 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.394 Å) | | Cite: | Structural basis of ubiquitin modification by the Legionella effector SdeA.
Nature, 557, 2018
|
|
