1DF5
 
 | |
4FC2
 
 | |
4F10
 
 | | Alginate lyase A1-III H192A complexed with tetrasaccharide | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, Alginate lyase, alpha-L-gulopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Mikami, B, Ban, M, Suzuki, S, Yoon, H.-J, Miyake, O, Yamasaki, M, Ogura, K, Maruyama, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2012-05-05 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced-fit motion of a lid loop involved in catalysis in alginate lyase A1-III
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EZB
 
 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
4F4N
 
 | |
1EAW
 
 | |
1EJQ
 
 | | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE | | Descriptor: | SYNDECAN-4 | | Authors: | Shin, J, Oh, E.S, Lee, D, Couchman, J.R, Lee, W. | | Deposit date: | 2000-03-04 | | Release date: | 2001-03-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE SYNDECAN-4 WHOLE CYTOPLASMIC DOMAIN IN THE PRESENCE OF PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE
To be Published
|
|
4F3X
 
 | | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Putative aldehyde dehydrogenase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-09 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of putative aldehyde dehydrogenase from Sinorhizobium meliloti 1021 complexed with NAD
To be Published
|
|
1E2A
 
 | | ENZYME IIA FROM THE LACTOSE SPECIFIC PTS FROM LACTOCOCCUS LACTIS | | Descriptor: | ENZYME IIA, MAGNESIUM ION | | Authors: | Sliz, P, Engelmann, R, Hengstenberg, W, Pai, E.F. | | Deposit date: | 1997-04-25 | | Release date: | 1998-04-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of enzyme IIAlactose from Lactococcus lactis reveals a new fold and points to possible interactions of a multicomponent system.
Structure, 5, 1997
|
|
4FYG
 
 | | Structural basis for substrate recognition by a novel Legionella phosphoinositide phosphatase | | Descriptor: | (2R)-3-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6S)-2,3,6-trihydroxy-4,5-bis(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, SidF, inhibitor of growth family, ... | | Authors: | Hsu, F.S, Zhu, W, Brennan, L, Tao, L, Luo, Z.Q, Mao, Y. | | Deposit date: | 2012-07-04 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.822 Å) | | Cite: | Structural basis for substrate recognition by a unique Legionella phosphoinositide phosphatase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1DQN
 
 | | CRYSTAL STRUCTURE OF GIARDIA GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH A TRANSITION STATE ANALOGUE | | Descriptor: | GUANINE PHOSPHORIBOSYLTRANSFERASE, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Shi, W, Munagala, N.R, Wang, C.C, Li, C.M, Tyler, P.C, Furneaux, R.H, Grubmeyer, C, Schramm, V.L, Almo, S.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Giardia lamblia guanine phosphoribosyltransferase at 1.75 A(,).
Biochemistry, 39, 2000
|
|
1DX7
 
 | | Light-harvesting complex 1 beta subunit from Rhodobacter sphaeroides | | Descriptor: | Light harvesting 1 b(B850b) polypeptide | | Authors: | Conroy, M.J, Westerhuis, W, Parkes-Loach, P.S, Loach, P.A, Hunter, C.N, Williamson, M.P. | | Deposit date: | 1999-12-21 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure of Rhodobacter Sphaeroides Lh1 B Reveals Two Helical Domains Separated by a Flexible Region: Structural Consequences for the Lh1 Complex
J.Mol.Biol., 298, 2000
|
|
5COR
 
 | |
4G1E
 
 | | Crystal structure of integrin alpha V beta 3 with coil-coiled tag. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dong, X, Mi, L, Zhu, J, Wang, W, Luo, B, Springer, T.A. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | AlphaV Beta3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4GBX
 
 | | Crystal structure of an immune complex at pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II histocompatibility antigen, DM alpha chain, ... | | Authors: | Sethi, D.K, Pos, W, Wucherpfennig, K.W. | | Deposit date: | 2012-07-28 | | Release date: | 2013-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the HLA-DM-HLA-DR1 Complex Defines Mechanisms for Rapid Peptide Selection.
Cell(Cambridge,Mass.), 151, 2012
|
|
4FVD
 
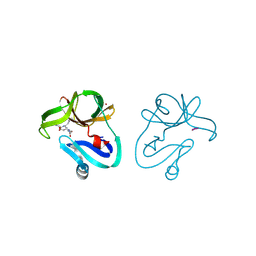 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
6JSU
 
 | |
4FVB
 
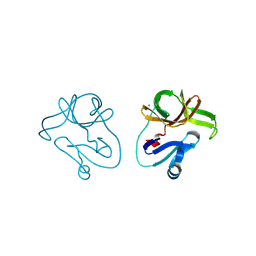 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FYP
 
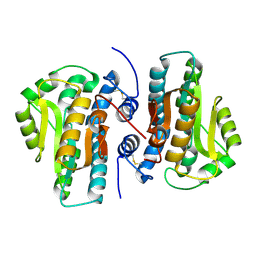 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
4G6H
 
 | | Crystal structure of NDH with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Li, W, Feng, Y, Ge, J, Yang, M. | | Deposit date: | 2012-07-19 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.262 Å) | | Cite: | Structural insight into the type-II mitochondrial NADH dehydrogenases.
Nature, 491, 2012
|
|
6JST
 
 | |
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
1CT2
 
 | | CRYSTAL STRUCTURE OF THE OMTKY3 P1 VARIANT OMTKY3-THR18I IN COMPLEX WITH SGPB | | Descriptor: | OVOMUCOID INHIBITOR, PROTEINASE B | | Authors: | Bateman, K.S, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N. | | Deposit date: | 1999-08-18 | | Release date: | 2000-01-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Deleterious effects of beta-branched residues in the S1 specificity pocket of Streptomyces griseus proteinase B (SGPB): crystal structures of the turkey ovomucoid third domain variants Ile18I, Val18I, Thr18I, and Ser18I in complex with SGPB.
Protein Sci., 9, 2000
|
|
1CZ4
 
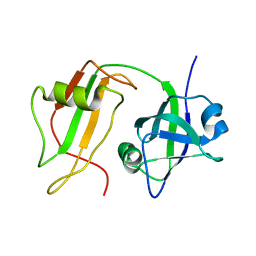 | | NMR STRUCTURE OF VAT-N: THE N-TERMINAL DOMAIN OF VAT (VCP-LIKE ATPASE OF THERMOPLASMA) | | Descriptor: | VCP-LIKE ATPASE | | Authors: | Coles, M, Diercks, T, Liermann, J, Groeger, A, Rockel, B, Baumeister, W, Koretke, K, Lupas, A, Peters, J, Kessler, H. | | Deposit date: | 1999-09-01 | | Release date: | 1999-10-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of VAT-N reveals a 'missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element.
Curr.Biol., 9, 1999
|
|
6KN5
 
 | |
